Releases: JohnSnowLabs/nlu
Databricks-Serve-Endpoint-Mode and Bug-Fixes in John Snow Labs NLU 5.0.1
- fix bug that caused predicted column names to change when saving/reloading a pipe
- fix bug causing some Visual based nlu components to use wrong data types
- New Databricks-Endpoint based inference mode. It is enabled if the env variable
DB_ENDPOINT_ENVis present. When enabled, the first row of every pandas dataframe passed topipe.predict()is checked for parameters. If your dataframe contains ofoutput_level,positions,keep_stranger_features,metadata,multithread,drop_irrelevant_cols,return_spark_df,get_embeddings, the first row of your dataframe is mapped to the corrosponding parameter and used to callpipeline.predict()
Medical Text Generation, ConvNext for image Classification and DistilBert,Bert,Roberta for Zero-Shot Classification in John Snow Labs NLU 5.0.0
We are very excited to announce NLU 5.0.0 has been released!
It comes with ZeroShotClassification models based on Bert, DistilBert, and Roberta architectures.
Additionally Medical Text Generator based on Bio-GPT as-well as a Bart based General Text Generator are now available in NLU.
Finally, ConvNextForImageClassification is an image classifier based on ConvNet models.
ConvNextForImageClassification
Tutorial Notebook
ConvNextForImageClassification is an image classifier based on ConvNet models.
The ConvNeXT model was proposed in A ConvNet for the 2020s by Zhuang Liu, Hanzi Mao, Chao-Yuan Wu, Christoph Feichtenhofer, Trevor Darrell, Saining Xie. ConvNeXT is a pure convolutional model (ConvNet), inspired by the design of Vision Transformers, that claims to outperform them.
Powered by ConvNextForImageClassification
Reference: A ConvNet for the 2020s
New NLU Models:
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.classify_image.convnext.tiny | image_classifier_convnext_tiny_224_local | Image Classification | ConvNextImageClassifier |
| en | en.classify_image.convnext.tiny | image_classifier_convnext_tiny_224_local | Image Classification | ConvNextImageClassifier |
DistilBertForZeroShotClassification
DistilBertForZeroShotClassification using a ModelForSequenceClassification trained on NLI (natural language inference) tasks.
Any combination of sequences and labels can be passed and each combination will be posed as a premise/hypothesis pair and passed to the pretrained model.
Powered by DistilBertForZeroShotClassification
New NLU Models:
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.distilbert.zero_shot_classifier | distilbert_base_zero_shot_classifier_uncased_mnli | Zero-Shot Classification | DistilBertForZeroShotClassification |
| tr | tr.distilbert.zero_shot_classifier.multinli | distilbert_base_zero_shot_classifier_turkish_cased_multinli | Zero-Shot Classification | DistilBertForZeroShotClassification |
| tr | tr.distilbert.zero_shot_classifier.allnli | distilbert_base_zero_shot_classifier_turkish_cased_allnli | Zero-Shot Classification | DistilBertForZeroShotClassification |
| tr | tr.distilbert.zero_shot_classifier.snli | distilbert_base_zero_shot_classifier_turkish_cased_snli | Zero-Shot Classification | DistilBertForZeroShotClassification |
BertForZeroShotClassification
Tutorial Notebook
BertForZeroShotClassification using a ModelForSequenceClassification trained on NLI (natural language inference) tasks.
Any combination of sequences and labels can be passed and each combination will be posed as a premise/hypothesis pair and passed to the pretrained model.
Powered by BertForZeroShotClassification
New NLU Models:
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.bert.zero_shot_classifier | bert_base_cased_zero_shot_classifier_xnli | Zero-Shot Classification | BertForZeroShotClassification |
RoBertaForZeroShotClassification
Tutorial Notebook
RoBertaForZeroShotClassification using a ModelForSequenceClassification trained on NLI (natural language inference) tasks.
Any combination of sequences and labels can be passed and each combination will be posed as a premise/hypothesis pair and passed to the pretrained model.
Powered by RoBertaForZeroShotClassification
New NLU Models:
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.roberta.zero_shot_classifier | roberta_base_zero_shot_classifier_nli | Zero-Shot Classification | RoBertaForZeroShotClassification |
BartTransformer
The Facebook BART (Bidirectional and Auto-Regressive Transformer) model is a state-of-the-art language generation model that was introduced by Facebook AI in 2019. It is based on the transformer architecture and is designed to handle a wide range of natural language processing tasks such as text generation, summarization, and machine translation.
BART is unique in that it is both bidirectional and auto-regressive, meaning that it can generate text both from left-to-right and from right-to-left. This allows it to capture contextual information from both past and future tokens in a sentence,resulting in more accurate and natural language generation.
The model was trained on a large corpus of text data using a combination of unsupervised and supervised learning techniques. It incorporates pretraining and fine-tuning phases, where the model is fi...
Medical Summarizer Models in John Snow Labs NLU 4.2.2
New Medical Summarizers:
- 'en.summarize.clinical_jsl'
- 'en.summarize.clinical_jsl_augmented'
- 'en.summarize.biomedical_pubmed'
- 'en.summarize.generic_jsl'
- 'en.summarize.clinical_questions'
- 'en.summarize.radiology'
- 'en.summarize.clinical_guidelines_large'
- 'en.summarize.clinical_laymen'
Hotfix Databricks save and reload models in John Snow Labs NLU 4.2.1
Bugfixes for saving and reloading pipelines on databricks
Support for Speech2Text, Images-Classification, Tabular Data, Zero-Shot-NER, via Wav2Vec2, Tapas, VIT , 4000+ New Models, 90+ Languages, in John Snow Labs NLU 4.2.0
Support for Speech2Text, Images-Classification, Tabular Data, Zero-Shot-NER, via Wav2Vec2, Tapas, VIT , 4000+ New Models, 90+ Languages, in John Snow Labs NLU 4.2.0
We are incredibly excited to announce NLU 4.2.0 has been released with new 4000+ models in 90+ languages and support for new 8 Deep Learning Architectures.
4 new tasks are included for the very first time,
Zero-Shot-NER, Automatic Speech Recognition, Image Classification and Table Question Answering powered
by Wav2Vec 2.0, HuBERT, TAPAS, VIT, SWIN, Zero-Shot-NER.
Additionally, CamemBERT based architectures are available for Sequence and Token Classification powered by Spark-NLPs
CamemBertForSequenceClassification and CamemBertForTokenClassification
Automatic Speech Recognition (ASR)
Demo Notebook
Wav2Vec 2.0 and HuBERT enable ASR for the very first time in NLU.
Wav2Vec2 is a transformer model for speech recognition that uses unsupervised pre-training on large amounts of unlabeled speech data to improve the accuracy of automatic speech recognition (ASR) systems. It is based on a self-supervised learning approach that learns to predict masked portions of speech signal, and has shown promising results in reducing the amount of labeled training data required for ASR tasks.
These Models are powered by Spark-NLP's Wav2Vec2ForCTC Annotator

HuBERT models match or surpass the SOTA approaches for speech representation learning for speech recognition, generation, and compression. The Hidden-Unit BERT (HuBERT) approach was proposed for self-supervised speech representation learning, which utilizes an offline clustering step to provide aligned target labels for a BERT-like prediction loss.
These Models is powered by Spark-NLP's HubertForCTC Annotator
Usage
You just need an audio-file on disk and pass the path to it or a folder of audio-files.
import nlu
# Let's download an audio file
!wget https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/audio/samples/wavs/ngm_12484_01067234848.wav
# Let's listen to it
from IPython.display import Audio
FILE_PATH = "ngm_12484_01067234848.wav"
asr_df = nlu.load('en.speech2text.wav2vec2.v2_base_960h').predict('ngm_12484_01067234848.wav')
asr_df| text |
|---|
| PEOPLE WHO DIED WHILE LIVING IN OTHER PLACES |
To test out HuBERT you just need to update the parameter for load()
asr_df = nlu.load('en.speech2text.hubert').predict('ngm_12484_01067234848.wav')
asr_dfImage Classification
For the first time ever NLU introduces state-of-the-art image classifiers based on
VIT and Swin giving you access to hundreds of image classifiers for various domains.
Inspired by the Transformer scaling successes in NLP, the researchers experimented with applying a standard Transformer directly to images, with the fewest possible modifications. To do so, images are split into patches and the sequence of linear embeddings of these patches were provided as an input to a Transformer. Image patches were actually treated the same way as tokens (words) in an NLP application. Image classification models were trained in supervised fashion.
You can check Scale Vision Transformers (ViT) Beyond Hugging Face article to learn deeper how ViT works and how it is implemeted in Spark NLP.
This is Powerd by Spark-NLP's VitForImageClassification Annotator
Swin is a hierarchical Transformer whose representation is computed with Shifted windows.
The shifted windowing scheme brings greater efficiency by limiting self-attention computation to non-overlapping local windows while also allowing for cross-window connection.
This hierarchical architecture has the flexibility to model at various scales and has linear computational complexity with respect to image size. These qualities of Swin Transformer make it compatible with a broad range of vision tasks
This is powerd by Spark-NLP's Swin For Image Classification
Swin Transformer: Hierarchical Vision Transformer using Shifted Windows by Ze Liu, Yutong Lin, Yue Cao, Han Hu, Yixuan Wei, Zheng Zhang, Stephen Lin, Baining Guo.
Usage:
# Download an image
os.system('wget https://raw.githubusercontent.com/JohnSnowLabs/nlu/release/4.2.0/tests/datasets/ocr/vit/ox.jpg')
# Load VIT model and predict on image file
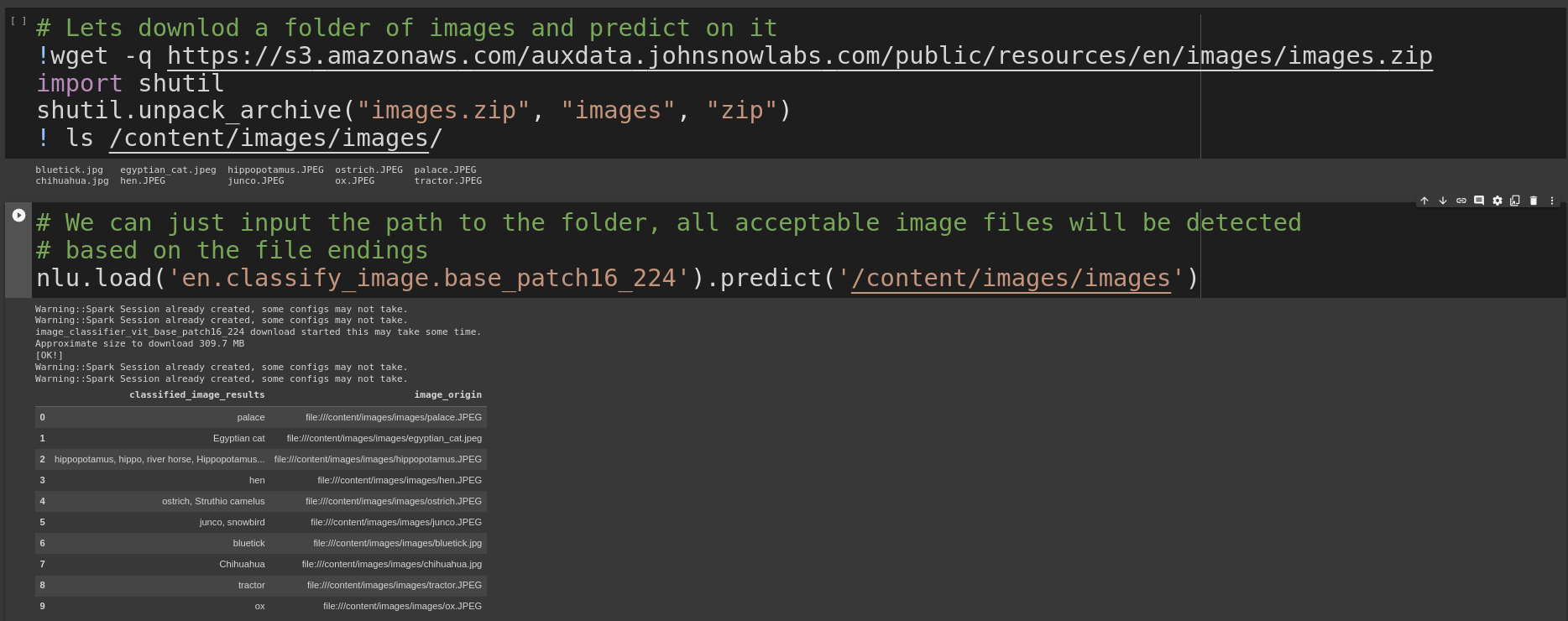
vit = nlu.load('en.classify_image.base_patch16_224').predict('ox.jpg')Lets download a folder of images and predict on it
!wget -q https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/images/images.zip
import shutil
shutil.unpack_archive("images.zip", "images", "zip")
! ls /content/images/images/Once we have image data its easy to label it, we just pass the folder with images to nlu.predict()
and NLU will return a pandas DF with one row per image detected
nlu.load('en.classify_image.base_patch16_224').predict('/content/images/images')To use SWIN we just update the parameter to load()
load('en.classify_image.swin.tiny').predict('/content/images/images')Visual Table Question Answering
TapasForQuestionAnswering can load TAPAS Models with a cell selection head and optional aggregation head on top for question-answering tasks on tables (linear layers on top of the hidden-states output to compute logits and optional logits_aggregation), e.g. for SQA, WTQ or WikiSQL-supervised tasks. TAPAS is a BERT-based model specifically designed (and pre-trained) for answering questions about tabular data.
Powered by TAPAS: Weakly Supervised Table Parsing via Pre-training

Usage:
First we need a pandas dataframe on for which we want to ask questions. The so called "context"
import pandas as pd
context_df = pd.DataFrame({
'name':['Donald Trump','Elon Musk'],
'money': ['$100,000,000','$20,000,000,000,000'],
'married': ['yes','no'],
'age' : ['75','55'] })
context_dfThen we create an array of questions
questions = [
"Who earns less than 200,000,000?",
"Who earns more than 200,000,000?",
"Who earns 100,000,000?",
"How much money has Donald Trump?",
"Who is the youngest?",
]
questionsNow Combine the data, pass it to NLU and get answers for your questions
import nlu
# Now we combine both to a tuple and we are done! We can now pass this to the .predict() method
tapas_data = (context_df, questions)
# Lets load a TAPAS QA model and predict on (context,question).
# It will give us an aswer for every question in the questions array, based on the context in context_df
answers = nlu.load('en.answer_question.tapas.wtq.large_finetuned').predict(tapas_data)
answers| sentence | tapas_qa_UNIQUE_aggregation | tapas_qa_UNIQUE_answer | tapas_qa_UNIQUE_cell_positions | tapas_qa_UNIQUE_cell_scores | tapas_qa_UNIQUE_origin_question |
|---|---|---|---|---|---|
| Who earns less than 200,000,000? | NONE | Donald Trump | [0, 0] | 1 | Who earns less than 200,000,000? |
| Who earns more than 200,000,000? | NONE | Elon Musk | [0, 1] | ... |
OCR Visual Tables into Pandas DataFrames from PDF/DOCX/PPT files, 1000+ new state-of-the-art transformer models for Question Answering (QA) for over 30 languages, up to 700% speedup on GPU, 20 Biomedical models for over 8 languages, 50+ Terminology Code Mappers between RXNORM, NDC, UMLS,ICD10, ICDO, UMLS, SNOMED and MESH, Deidentification in Romanian, various Spark NLP helper methods and much more in 1 line of code with John Snow Labs NLU 4.0.0
OCR Visual Tables into Pandas DataFrames from PDF/DOC(X)/PPT files, 1000+ new state-of-the-art transformer models for Question Answering (QA) for over 30 languages, up to 700% speedup on GPU, 20 Biomedical models for over 8 languages, 50+ Terminology Code Mappers between RXNORM, NDC, UMLS,ICD10, ICDO, UMLS, SNOMED and MESH, Deidentification in Romanian, various Spark NLP helper methods and much more in 1 line of code with John Snow Labs NLU 4.0.0
NLU 4.0 for OCR Overview
On the OCR side, we now support extracting tables from PDF/DOC(X)/PPT files into structured pandas dataframe, making it easier than ever before to analyze bulks of files visually!
Checkout the OCR Tutorial for extracting Tables from Image/PDF/DOC(X) files to see this in action
These models grab all Table data from the files detected and return a list of Pandas DataFrames,
containing Pandas DataFrame for every table detected
| NLU Spell | Transformer Class |
|---|---|
nlu.load(pdf2table) |
PdfToTextTable |
nlu.load(ppt2table) |
PptToTextTable |
nlu.load(doc2table) |
DocToTextTable |
This is powerd by John Snow Labs Spark OCR Annotataors for PdfToTextTable, DocToTextTable, PptToTextTable
NLU 4.0 Core Overview
-
On the NLU core side we have over 1000+ new state-of-the-art models in over 30 languages for modern extractive transformer-based Question Answering problems powerd by the ALBERT/BERT/DistilBERT/DeBERTa/RoBERTa/Longformer Spark NLP Annotators trained on various SQUAD-like QA datasets for domains like Twitter, Tech, News, Biomedical COVID-19 and in various model subflavors like sci_bert, electra, mini_lm, covid_bert, bio_bert, indo_bert, muril, sapbert, bioformer, link_bert, mac_bert
-
Additionally up to 700% speedup transformer-based Word Embeddings on GPU and up to 97% speedup on CPU for tensorflow operations, support for Apple M1 chips, Pyspark 3.2 and 3.3 support.
Ontop of this, we are now supporting Apple M1 based architectures and every Pyspark 3.X version, while deprecating support for Pyspark 2.X. -
Finally, NLU-Core features various new helper methods for working with Spark NLP and embellishes now the entire universe of Annotators defined by Spark NLP and Spark NLP for healthcare.
NLU 4.0 for Healthcare Overview
-
On the healthcare side NLU features 20 Biomedical models for over 8 languages (English, French, Italian, Portuguese, Romanian, Catalan and Galician) detect entities like
HUMANandSPECIESbased on LivingNER corpus -
Romanian models for Deidentification and extracting Medical entities like
Measurements,Form,Symptom,Route,Procedure,Disease_Syndrome_Disorder,Score,Drug_Ingredient,Pulse,Frequency,Date,Body_Part,Drug_Brand_Name,Time,Direction,Dosage,Medical_Device,Imaging_Technique,Test,Imaging_Findings,Imaging_Test,Test_Result,Weight,Clinical_DeptandUnitswith SPELL and SPELL respectively -
English NER models for parsing entities in Clinical Trial Abstracts like
Age,AllocationRatio,Author,BioAndMedicalUnit,CTAnalysisApproach,CTDesign,Confidence,Country,DisorderOrSyndrome,DoseValue,Drug,DrugTime,Duration,Journal,NumberPatients,PMID,PValue,PercentagePatients,PublicationYear,TimePoint,Valueusingen.med_ner.clinical_trials_abstracts.pipeand also Pathogen NER models forPathogen,MedicalCondition,Medicinewithen.med_ner.pathogenandGENE_PROTEINwithen.med_ner.biomedical_bc2gm.pipeline -
First Public Health Model for Emotional Stress classification It is a PHS-BERT-based model and trained with the Dreaddit dataset using
en.classify.stress -
50 + new Entity Mappers for problems like :
- Extract section headers in scientific articles and normalize them with
en.map_entity.section_headers_normalized - Map medical abbreviates to their definitions with
en.map_entity.abbreviation_to_definition - Map drugs to action and treatments with
en.map_entity.drug_to_action_treatment - Map drug brand to their National Drug Code (NDC) with
en.map_entity.drug_brand_to_ndc - Convert between terminologies using
en.<START_TERMINOLOGY>_to_<TARGET_TERMINOLOGY>- This works for the terminologies
rxnorm,ndc,umls,icd10cm,icdo,umls,snomed,meshsnomed_to_icdosnomed_to_icd10cmrxnorm_to_umls
- This works for the terminologies
- powerd by Spark NLP for Healthcares ChunkMapper Annotator
- Extract section headers in scientific articles and normalize them with
Extract Tables from PDF files as Pandas DataFrames
nlu.load('pdf2table').predict('/path/to/sample.pdf') Output of PDF Table OCR :
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear |
|---|---|---|---|---|---|---|---|---|---|
| 21 | 6 | 160 | 110 | 3.9 | 2.62 | 16.46 | 0 | 1 | 4 |
| 21 | 6 | 160 | 110 | 3.9 | 2.875 | 17.02 | 0 | 1 | 4 |
| 22.8 | 4 | 108 | 93 | 3.85 | 2.32 | 18.61 | 1 | 1 | 4 |
| 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 |
| 18.7 | 8 | 360 | 175 | 3.15 | 3.44 | 17.02 | 0 | 0 | 3 |
| 13.3 | 8 | 350 | 245 | 3.73 | 3.84 | 15.41 | 0 | 0 | 3 |
| 19.2 | 8 | 400 | 175 | 3.08 | 3.845 | 17.05 | 0 | 0 | 3 |
| 27.3 | 4 | 79 | 66 | 4.08 | 1.935 | 18.9 | 1 | 1 | 4 |
| 26 | 4 | 120.3 | 91 | 4.43 | 2.14 | 16.7 | 0 | 1 | 5 |
| 30.4 | 4 | 95.1 | 113 | 3.77 | 1.513 | 16.9 | 1 | 1 | 5 |
| 15.8 | 8 | 351 | 264 | 4.22 | 3.17 | 14.5 | 0 | 1 | 5 |
| 19.7 | 6 | 145 | 175 | 3.62 | 2.77 | 15.5 | 0 | 1 | 5 |
| 15 | 8 | 301 | 335 | 3.54 | 3.57 | 14.6 | 0 | 1 | 5 |
| 21.4 | 4 | 121 | 109 | 4.11 | 2.78 | 18.6 | 1 | 1 | 4 |
Extract Tables from DOC/DOCX files as Pandas DataFrames
nlu.load('doc2table').predict('/path/to/sample.docx') Output of DOCX Table OCR :
| Screen Reader | Responses | Share |
|---|---|---|
| JAWS | 853 | 49% |
| NVDA | 238 | 14% |
| Window-Eyes | 214 | 12% |
| System Access | 181 | 10% |
| VoiceOver | 159 | 9% |
Extract Tables from PPT files as Pandas DataFrame
nlu.load('ppt2table').predict('/path/to/sample.docx') Output of PPT Table OCR :
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
and
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 6.7 | 3.3 | 5.7 | 2.5 | virginica |
| ... |
600 new models for over 75 new languages including Ancient, Dead and Extinct languages, 155 languages total covered, 400% Tokenizer Speedup, 18x USE-Embeddings GPU speedup in John Snow Labs NLU 3.4.4
We are very excited to announce NLU 3.4.4 has been released with over 600 new models, over 75 new languages, and 155 languages covered in total,
400% speedup for tokenizers and 18x speedup of UniversalSentenceEncoder on GPU.
On the general NLP side, we have transformer-based Embeddings and Token Classifiers powered by state of the art CamemBertEmbeddings and DeBertaForTokenClassification based
architectures as well as various new models for
Historical, Ancient,Dead, Extinct, Genetic, and Constructed languages like
Old Church Slavonic, Latin, Sanskrit, Esperanto, Volapük, Coptic, Nahuatl, Ancient Greek (to 1453), Old Russian.
On the healthcare side, we have Portuguese De-identification Models, have NER models for Gene detection and finally RxNorm Sentence resolution model for mapping and extracting pharmaceutical actions (e.g. analgesic, hypoglycemic)
as well as treatments (e.g. backache, diabetes).
For full release notes with all models see
here
or here ,
First-time language models covered
The languages for these models are covered for the very first time ever by NLU.
| Number | Language Name(s) | NLU Reference | Spark NLP Reference | Task | Annotator Class | Scope | Language Type |
|---|---|---|---|---|---|---|---|
| 0 | Sanskrit | sa.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Ancient |
| 1 | Sanskrit | sa.lemma | lemma_vedic | Lemmatization | LemmatizerModel | Individual | Ancient |
| 2 | Sanskrit | sa.pos | pos_vedic | Part of Speech Tagging | PerceptronModel | Individual | Ancient |
| 3 | Sanskrit | sa.stopwords | stopwords_iso | Stop Words Removal | StopWordsCleaner | Individual | Ancient |
| 4 | Volapük | vo.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Constructed |
| 5 | Nahuatl languages | nah.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Collective | Genetic |
| 6 | Aragonese | an.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 7 | Assamese | as.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 8 | Asturian, Asturleonese, Bable, Leonese | ast.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 9 | Bashkir | ba.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 10 | Bavarian | bar.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 11 | Bishnupriya | bpy.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 12 | Burmese | my.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 13 | Cebuano | ceb.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 14 | Central Bikol | bcl.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 15 | Chechen | ce.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 16 | Chuvash | cv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 17 | Corsican | co.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 18 | Dhivehi, Divehi, Maldivian | dv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 19 | Egyptian Arabic | arz.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 20 | Emiliano-Romagnolo | eml.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 21 | Erzya | myv.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 22 | Georgian | ka.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 23 | Goan Konkani | gom.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel | Individual | Living |
| 24 | Javanese | jv.embed.distilbert | distilbert_embeddings_javanese_distilbert_small | Embeddings | DistilBertEmbeddings | Individual | Living |
| 25 | Javanese | jv.embed.javanese_distilbert_small_imdb | distilbert_embeddings_javanese_distilbert_small_imdb | Embeddings | DistilBertEmbeddings | Individual | Living |
| 26 | Javanese | jv.embed.javanese_roberta_small | roberta_embeddings_javanese_roberta_small | Embeddings | RoBertaEmbeddings | Individual | Living |
| 27 | Javanese | [jv.embed.javanese_roberta_small_imdb](https://nlp.jo... |
Zero-Shot-Relation-Extraction, DeBERTa for Sequence Classification, 150+ new models, 60+ Languages in John Snow Labs NLU 3.4.3
We are very excited to announce NLU 3.4.3 has been released!
This release features new models for Zero-Shot-Relation-Extraction, DeBERTa for Sequence Classification,
Deidentification in French and Italian and
Lemmatizers, Parts of Speech Taggers, and Word2Vec Embeddings for over 66 languages, with 20 languages being covered
for the first time by NLU, including ancient and exotic languages like Ancient Greek, Old Russian,
Old French and much more. Once again we would like to thank our community to make this release possible.
NLU for Healthcare
On the healthcare NLP side, a new ZeroShotRelationExtractionModel is available, which can extract relations between
clinical entities in an unsupervised fashion, no training required!
Additionally, New French and Italian Deidentification models are available for clinical and healthcare domains.
Powerd by the fantastic Spark NLP for helathcare 3.5.0 release
Zero-Shot Relation Extraction
Zero-shot Relation Extraction to extract relations between clinical entities with no training dataset
import nlu
pipe = nlu.load('med_ner.clinical relation.zeroshot_biobert')
# Configure relations to extract
pipe['zero_shot_relation_extraction'].setRelationalCategories({
"CURE": ["{{TREATMENT}} cures {{PROBLEM}}."],
"IMPROVE": ["{{TREATMENT}} improves {{PROBLEM}}.", "{{TREATMENT}} cures {{PROBLEM}}."],
"REVEAL": ["{{TEST}} reveals {{PROBLEM}}."]})
.setMultiLabel(False)
df = pipe.predict("Paracetamol can alleviate headache or sickness. An MRI test can be used to find cancer.")
df[
'relation', 'relation_confidence', 'relation_entity1', 'relation_entity1_class', 'relation_entity2', 'relation_entity2_class',]
# Results in following table :| relation | relation_confidence | relation_entity1 | relation_entity1_class | relation_entity2 | relation_entity2_class |
|---|---|---|---|---|---|
| REVEAL | 0.976004 | An MRI test | TEST | cancer | PROBLEM |
| IMPROVE | 0.988195 | Paracetamol | TREATMENT | sickness | PROBLEM |
| IMPROVE | 0.992962 | Paracetamol | TREATMENT | headache | PROBLEM |
New Healthcare Models overview
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.relation.zeroshot_biobert | re_zeroshot_biobert | Relation Extraction | ZeroShotRelationExtractionModel |
| fr | fr.med_ner.deid_generic | ner_deid_generic | De-identification | MedicalNerModel |
| fr | fr.med_ner.deid_subentity | ner_deid_subentity | De-identification | MedicalNerModel |
| it | it.med_ner.deid_generic | ner_deid_generic | Named Entity Recognition | MedicalNerModel |
| it | it.med_ner.deid_subentity | ner_deid_subentity | Named Entity Recognition | MedicalNerModel |
NLU general
On the general NLP side we have new transformer based DeBERTa v3 sequence classifiers models fine-tuned in Urdu, French and English for
Sentiment and News classification. Additionally, 100+ Part Of Speech Taggers and Lemmatizers for 66 Languages and for 7
languages new word2vec embeddings, including hi,azb,bo,diq,cy,es,it,
powered by the amazing Spark NLP 3.4.3 release
New Languages covered:
First time languages covered by NLU are :
South Azerbaijani, Tibetan, Dimli, Central Kurdish, Southern Altai,
Scottish Gaelic,Faroese,Literary Chinese,Ancient Greek,
Gothic, Old Russian, Church Slavic,
Old French,Uighur,Coptic,Croatian, Belarusian, Serbian
and their respective ISO-639-3 and ISO 630-2 codes are :
azb,bo,diq,ckb, lt gd, fo,lzh,grc,got,orv,cu,fro,qtd,ug,cop,hr,be,qhe,sr
New NLP Models Overview
Multilingual DeBERTa Transformer Embeddings for 100+ Languages, Spanish Deidentification and NER for Randomized Clinical Trials - John Snow Labs NLU 3.4.2
Multilingual DeBERTa Transformer Embeddings for 100+ Languages, Spanish Deidentification and NER for Randomized Clinical Trials - John Snow Labs NLU 3.4.2
We are very excited NLU 3.4.2 has been released.
On the open source side we have 5 new DeBERTa Transformer models for English and Multi-Lingual for 100+ languages.
DeBERTa improves over BERT and RoBERTa by introducing two novel techniques.
For the healthcare side we have new NER models for randomized clinical trials (RCT) which can detect entities of type
BACKGROUND, CONCLUSIONS, METHODS, OBJECTIVE, RESULTS from clinical text.
Additionally, new Spanish Deidentification NER models for entities like STATE, PATIENT, DEVICE, COUNTRY, ZIP, PHONE, HOSPITAL and many more.
New Open Source Models
Integrates models from Spark NLP 3.4.2 release
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.embed.deberta_v3_xsmall | deberta_v3_xsmall | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_small | deberta_v3_small | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_base | deberta_v3_base | Embeddings | DeBertaEmbeddings |
| en | en.embed.deberta_v3_large | deberta_v3_large | Embeddings | DeBertaEmbeddings |
| xx | xx.embed.mdeberta_v3_base | mdeberta_v3_base | Embeddings | DeBertaEmbeddings |
New Healthcare Models
Integrates models from Spark NLP For Healthcare 3.4.2 release
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| en | en.med_ner.clinical_trials | bert_sequence_classifier_rct_biobert | Text Classification | MedicalBertForSequenceClassification |
| es | es.med_ner.deid.generic.roberta | ner_deid_generic_roberta_augmented | De-identification | MedicalNerModel |
| es | es.med_ner.deid.subentity.roberta | ner_deid_subentity_roberta_augmented | De-identification | MedicalNerModel |
| en | en.med_ner.deid.generic_augmented | ner_deid_generic_augmented | ['Named Entity Recognition', 'De-identification'] | MedicalNerModel |
| en | en.med_ner.deid.subentity_augmented | ner_deid_subentity_augmented | ['Named Entity Recognition', 'De-identification'] | MedicalNerModel |
Additional NLU resources
- 140+ NLU Tutorials
- NLU in Action
- Streamlit visualizations docs
- The complete list of all 4000+ models & pipelines in 200+ languages is available on Models Hub.
- Spark NLP publications
- NLU documentation
- Discussions Engage with other community members, share ideas, and show off how you use Spark NLP and NLU!
1 line Install NLU on Google Colab
!wget https://setup.johnsnowlabs.com/nlu/colab.sh -O - | bash
1 line Install NLU on Kaggle
!wget https://setup.johnsnowlabs.com/nlu/kaggle.sh -O - | bash
Install via PIP
! pip install nlu pyspark streamlit==0.80.0
22 New models for 23 languages including various African and Indian languages, Medical Spanish models and more in NLU 3.4.1
We are very excited to announce the release of NLU 3.4.1
which features 22 new models for 23 languages where the
The open-source side covers new Embeddings for Vietnamese and English Clinical domains and Multilingual Embeddings for 12 Indian and 9 African Languages.
Additionally, there are new Sequence classifiers for Multilingual NER for 9 African languages,
German Sentiment Classifiers and English Emotion and Typo Classifiers.
The healthcare side covers Medical Spanish models, Classifiers for Drugs, Gender, the Pico Framework, and Relation Extractors for Adverse Drug events and Temporality.
Finally, Spark 3.2.X is now supported and bugs related to Databricks environments have been fixed.
General NLU Improvements
- Support for Spark 3.2.x
New Open Source Models
Based on the amazing 3.4.1 Spark NLP Release
integrates new Multilingual embeddings for 12 Major Indian languages,
embeddings for Vietnamese, French, and English Clinical domains.
Additionally new Multilingual NER model for 9 African languages, English 6 Class Emotion classifier and Typo detectors.
New Embeddings
- Multilingual ALBERT - IndicBert model pretrained exclusively on 12 major Indian languages with size smaller and performance on par or better than competing models. Languages covered are Assamese, Bengali, English, Gujarati, Hindi, Kannada, Malayalam, Marathi, Oriya, Punjabi, Tamil, Telugu.
Available with xx.embed.albert.indic - Fine tuned Vietnamese DistilBERT Base cased embeddings. Available with vi.embed.distilbert.cased
- Clinical Longformer Embeddings which consistently out-performs ClinicalBERT for various downstream
tasks and on datasets. Available with en.embed.longformer.clinical - Fine tuned Static French Word2Vec Embeddings in 3 sizes, 200d, 300d and 100d. Available with fr.embed.word2vec_wiki_1000, fr.embed.word2vec_wac_200 and fr.embed.w2v_cc_300d
New Transformer based Token and Sequence Classifiers
- Multilingual NER Distilbert model which detects entities
DATE,LOC,ORG,PERfor the languages 9 African languages (Hausa, Igbo, Kinyarwanda, Luganda, Nigerian, Pidgin, Swahili, Wolof, and Yorùbá).
Available with xx.ner.masakhaner.distilbert - German News Sentiment Classifier available with de.classify.news_sentiment.bert
- English Emotion Classifier for 6 Classes available with en.classify.emotion.bert
- **English Typo Detector **: available with en.classify.typos.distilbert
| Language | NLU Reference | Spark NLP Reference | Task | Annotator Class |
|---|---|---|---|---|
| xx | xx.embed.albert.indic | albert_indic | Embeddings | AlbertEmbeddings |
| xx | xx.ner.masakhaner.distilbert | xlm_roberta_large_token_classifier_masakhaner | Named Entity Recognition | DistilBertForTokenClassification |
| en | en.embed.longformer.clinical | clinical_longformer | Embeddings | LongformerEmbeddings |
| en | en.classify.emotion.bert | bert_sequence_classifier_emotion | Text Classification | BertForSequenceClassification |
| de | de.classify.news_sentiment.bert | bert_sequence_classifier_news_sentiment | Sentiment Analysis | BertForSequenceClassification |
| en | en.classify.typos.distilbert | distilbert_token_classifier_typo_detector | Named Entity Recognition | DistilBertForTokenClassification |
| fr | fr.embed.word2vec_wiki_1000 | word2vec_wiki_1000 | Embeddings | WordEmbeddingsModel |
| fr | fr.embed.word2vec_wac_200 | word2vec_wac_200 | Embeddings | WordEmbeddingsModel |
| fr | fr.embed.w2v_cc_300d | w2v_cc_300d | Embeddings | WordEmbeddingsModel |
| vi | vi.embed.distilbert.cased | distilbert_base_cased | Embeddings | DistilBertEmbeddings |
New Healthcare Models
Integrated from the amazing 3.4.1 Spark NLP For Healthcare Release.
which makes 2 new Annotator Classes available, MedicalBertForSequenceClassification and MedicalDistilBertForSequenceClassification,
various medical Spanish models, RxNorm Resolvers,
Transformer based sequence classifiers for Drugs, Gender and the PICO framework,
and Relation extractors for Temporality and Causality of Drugs and Adverse Events.
New Medical Spanish Models
- Spanish Word2Vec Embeddings available with es.embed.sciwiki_300d
- Spanish PHI Deidentification NER models with two different subsets of entities extracted, available with ner_deid_generic and ner_deid_subentity
New Resolvers
- RxNorm resolvers with augmented concept data available with en.med_ner.supplement_clinical
New Transformer based Sequence Classifiers
- Adverse Drug Event Classifier Biobert based available with en.classify.ade.seq_biobert
- Patient Gender Classifier Biobert and Distilbert based available with en.classify.gender.seq_biobert
and available with en.classify.ade.seq_distilbert - PiCO Framework Classifier available with en.classify.pico.seq_biobert
New Relation Extractors
- Temporal Relation Extractor available with en.relation.temporal_events_clinical
- Adverse Drug Event Relation Extractors one version Biobert Embeddings and one non-DL version available with en.relation.adverse_drug_events.clinical available with [en.relation.adverse_drug_events.clinical.biobert](https://nlp.johnsnowlabs.com/2021/07/12/redl_ade_biobert_en.html...