\n",
+ "Note: It is important to remember that the gaps do actually exist in reality and this is a caveat which must be considered when interpreting the transect derived from CTD casts. Indeed, if you look at the x-axis of the plot below you will see that the deployments are not necessarily regularly spaced and some gaps are larger than others.\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 32,
+ "id": "fcf8a137",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "image/png": "iVBORw0KGgoAAAANSUhEUgAAA3wAAAI2CAYAAAALoIqcAAAAOnRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjEwLjUsIGh0dHBzOi8vbWF0cGxvdGxpYi5vcmcvWftoOwAAAAlwSFlzAAAOxAAADsQBlSsOGwAAcO1JREFUeJzt3XlcVPX+x/H3gIAgKsmq4oZbi2saaosbpqZdQ6+lleWWS1rX0t9NTcvUXFrVMssyrW5aWllatFm4FYZ6XdPcpTQlkFJTEAfm+/vDnCsBCuOMMwyv5+9xHr/mfM98zvecO8J8+Jzv92sxxhgBAAAAALyOj7s7AAAAAABwDRI+AAAAAPBSJHwAAAAA4KVI+AAAAADAS5HwAQAAAICXIuEDAAAAAC9FwgcAAAAAXqqMuzvgLWw2m44fP66yZcvKYrG4uzsAAABAgYwxOnPmjEJCQuTj4/n1H6vVqpycHJeeo0yZMvLz83PpOdyFhM9Jjh8/rtDQUHd3AwAAACiSjIwMVapUyd3duCir1arqNSsq9UiWS89TpUoVpaSkeGXSR8LnJGXLlpUkHZ1+kwL9fN3cG8/hE+Pav8aUSBSAC5fu7g6UXOYUP86LwqcCP5Psyht398DtcsO974udI6zhQe7ugkc4Va2iu7twxZzJytE1UR/Zv796spycHKUeydJPqT1VNtA137HPZOXqmqgPlZOTQ8KHwp1/jDPQz1eB/iR85/kE8IUiHxK+wvm7uwMll+HnTpH4+PMzyY6fz8oN4N+NJJUpy32QpNzA0ve1uCQNQwos66NAF31WLca7fx6Wvk+2q9l8zm04hz+m5+d9fzhyntyS84vH09iy+cJWFJZMm7u74DmyvfsLTpEwEkOSFLDrpLu74BEO7Grn7i5cMWeyrZIWu7sbJdbSpUv1yiuvaOPGjTp58qSsVqvKlPlfWrVnzx4NGTJEP/zwgyIjI/Xkk09qwIABbusvmQkAAAAAj2axGZduxZGZman27dtrzJgx+dqsVqu6du2qsLAwbdiwQU888YSGDBmib7/91lm3otio8AEAAABAEfXp00eStGrVqnxtX3zxhQ4dOqRNmzapfPnyatCggVavXq2XX35ZcXFxV7in55DwOZmxndtwjiWHR4by4ZYUKvc0P5IclX2ynLu7UCL457p2lreSJCcrwN1dcLuDUW3c3QUARWUz5zZXxXaS9evX64YbblD58uXt++Li4gqsBl4pfLsCAAAAUOplZeX9o6Aja/OlpaUpIiIiz77w8HClp7tvKnLG8AEAAADwaBZjXLpJUmhoqIKCguzblClTit1P44EzflLhAwAAAFDqZWRkKDAw0P76wpk3iyoyMlK7du3Ksy89PV3h4eGX3T9HkfA5mbH5yORSOLWzursDHogxnoWynWHNCkedzQ689EGQMSz9cR6fGQAliSOzaRYntiQFBgbmSfgcERsbqxdeeEGnTp1ScHCwJCkxMVEtWrS47H46ioQPAAAAAIro999/1y+//KJ9+/ZJkrZu3SpfX1/VqVNHnTt3VtWqVTVgwABNmDBBycnJeu+99/TFF1+4rb8kfAAAAAA8msUmF1b4inf88uXL1b9/f/vr5s2bS5JWrlyptm3bKiEhQUOGDFGzZs0UGRmpV1991W1LMkgkfM5nLOc2nJPr7g6gJLHl+Lq7CyWWNYfHYYvCwjPVdtlnWZYBABzRr18/9evXr9D2+vXrF7hGn7uQ8AEAAADwbCVkHT5PxOwiAAAAAOClqPA5mcn1kfEhj7bL5fHWfCze/Veky8Lj0A6z2fi5UxTWXB59PS/HxlcAACXHhevluSK2N+MbAgAAAAB4Kf68BwAAAMCjXYl1+LwVCZ+TGWNhYd8LmBx398DzWCx8PgrDvx3Hce+KxmZjJtjzjPjMAEBpQMIHAAAAwLMxS6fDGMMHAAAAAF6KCp+zsfB6XszSmZ+fd/8V6XIYG58XuBazmf7PqQ713d0FACgyi3HdROfePoE6v/kAAAAAwEtR4QMAAADg0Zil03EkfE5my/WRjYXX/4dHOvPz8sU9LwuPQzvsj/YN3N0FAADggUj4AAAAAHg2Zul0GKUoAAAAAPBSVPicjVk68zA80pmfzd0d8FwHr2/j7i4AAAAPZDEuHMPn5cNtqPABAAAAgJeiwgcAAADAsxnjuonvvLzCR8LnZDabDwv7XoBHOvOz8EgnAAAArhASPgAAAAAejXX4HEcpCgAAAAC8FBU+JzPGIsMsnXYmh78p5MPnAwAAoHhYh89hfBsHAAAAAC9FhQ8AAACAR7OYc5urYnszKnwAAAAA4KWo8DmZMT4yhjz6PJPLvQAAAMBlYgyfw/g2DgAAAABeigofAAAAAI/GOnyOI+FzMpPrI+ND4fQ8Y+Ne5GNzdwcAAABQWpDwAQAAAPBsxpzbXBXbi1F+AQAAAAAvRYXPyYyxyBiLu7vhMUyOr7u74Hm8+49IAAAATscYPsdR4QMAAAAAL0WFDwAAAIBns8l1E995+YR6JHxOdm48KY90nmdj4fX8bHw+AAAAisNijCwumlzFVXE9Bd/GAQAAAMBLUeEDAAAA4Nls5tzmqthejITPyYx8ZCic2rHwen5e/tQAAAAAPAgJHwAAAADPxqQtDqP8AgAAAABeigqfk+XafJTLY4x2Nu4FAAAALhOzdDqOb+MAAAAA4KWo8AEAAADwbMzS6TASPiczxsLC6xew2Xzd3QXPw8LrAAAAuEK84pHON954QzfeeKMqVqyo8PBw/fOf/9SBAwfyHJOamqr4+HgFBQWpcuXKmjp1ar44CxYsUExMjAIDA9WmTRvt2bPnSl0CAAAAgMLYXLx5Ma9I+FavXq2+fftq7dq1+vbbb3XmzBnddtttslqt9mN69eql33//XUlJSZozZ46mTZum+fPn29sTExM1ePBgjR07Vhs2bFBUVJS6du2qs2fPuuOSAAAAAOCyWYzxvmlpjh49qipVqmjr1q1q1KiRtm3bpsaNG2v37t2qV6+eJOnJJ5/U8uXLtWXLFklSjx49FBgYqIULF0qSTp8+rfDwcC1atEjx8fGXPGdWVpaCgoK0/5G7FejHk7LnBfidcXcXPE756HR3d8Fj7ap+u7u7AACA1zuTbVXsP8cqMzNTgYGB7u7ORZ3/jn18VRsFlnXNUKGsM7kKabu6RNwPR3hFhe/vjh07JkmqVKmSJGn9+vWKjo62J3uSFBcXp+3btysrK8t+TPv27e3t5cqVU4sWLZScnFzgOaxWq7KysvJsAAAAAOBJvC7hM8Zo/Pjx6tSpk6KjoyVJaWlpioiIyHNceHi4bDabPTks7Ji0tLQCzzNlyhQFBQXZt9DQUBdcDQAAAAAZ87+ZOp29ed8Dj3l49LOHQ4cO1dy5cwttb9OmjVatWpVn36hRo7R9+3Z9//339n2ueGp13LhxGj16tP11VlaWQkNDZbP5sNj4BWyGWToBAAAAd/HohG/69OkaP358oe0BAQF5Xj/++ONasmSJ1q5dq8qVK9v3R0ZG5qvUpaeny8fHR2FhYZKkiIiIAo+pXbt2gef28/OTn59fsa4HAAAAgANcOZuml8/S6dEJX0hIiEJCQop07MSJEzVv3jytXr1atWrVytMWGxurw4cPa+/evapbt66kc7NyNmzY0D4wMzY2VitXrtTAgQMlSZmZmUpOTtaIESOcd0EAAAAAcAV5dMJXVNOnT9czzzyjpUuX6qqrrlJqaqqkc5O2+Pv7q1GjRmrdurUGDRqkWbNmKSUlRTNmzNDMmTPtMYYPH67OnTurXbt2atmypSZPnqwqVaqoS5cubroqAAAAAJJkMUYWF421c1VcT+EVCd9rr72mrKws3XbbbXn2r1y5Um3btpUkLV68WEOGDFGrVq1UoUIFjR49WgMGDLAfGxcXp7lz52rSpElKTU1VixYtlJCQIH9//2L1xRiLjLFc9jV5i1zGMwIAAABu4xUJX0pKyiWPiYqK0rJlyy56zIABA/IkgQAAAAA8AGP4HEb5BQAAAAC8lFdU+DyJTb7KZSkCO18b9yIfHvkFAAAonvNr5rkqthejwgcAAAAAXooKHwAAAADPZv7aXBXbi5HwOZnN+MhmKJyeZ2OWTgAAAMBtSPgAAAAAeDSLzcjiorF2rorrKSi/AAAAAICXosLnZLk2X+UyM6Vdrg/3AgAAAJeJMXwOo8IHAAAAAF6KCh8AAAAAz2b7a3NVbC9GwudkucZHuczSaccsnQAAAID7kPABAAAA8Gw2c25zVWwvRvkFAAAAALwUFT4ny7H5KodZOu3K+PA3hb/bVaOru7sAAABQsjBLp8P4Ng4AAAAAXooKHwAAAADPxiydDiPhczJm6czLZni8FQAAAHAXEj4AAAAAno0Kn8MoRQEAAACAl6LC52Q80pkXM5YCAADgclmMkcW4ZjpNV8X1FGQmAAAAAOClqPABAAAA8GyM4XMYFT4AAAAA8FJU+Jwsx+bDuLUL5Fpy3d0FAAAAlHTmr81Vsb0YFT4AAAAA8FJU+AAAAAB4NsbwOYyEz8lYliEv7kV+3BEAAABcKXz3BAAAAODZjP5X5XP25sAYvuPHj2vgwIGKiopScHCwbrzxRq1Zs+YyLtB1SPgAAAAAoBhGjhypDRs26JNPPtHWrVsVGxur22+/XX/88Ye7u5YPj3Q6Wa7xVY5hls7zuBf5+bu7AwAAACWNh83SmZycrEGDBqlly5aSpMmTJ2vWrFnavXu3fZ+noMIHAAAAoNTLysrKs1mt1kKPbdWqlZYtW6Zjx44pNzdX8+fPV5UqVdSgQYMr2OOiIeEDAAAA4NlcNX7vgtk/Q0NDFRQUZN+mTJlSaHdefvllhYWFKTw8XAEBAZo2bZoSEhIUHBzs7Cu/bDzS6WRnbb6ysPC6nb8Pf1MAAACA58vIyFBgYKD9dZkyhadKs2bN0t69e7VixQqFhobqnXfeUbdu3bR582aFhoZeie4WGQkfAAAAAM92BcbwBQYG5kn4CpOVlaUnn3xS33zzjVq3bi1Jatq0qRISErRo0SI9/PDDLuqoYyi/AAAAAEARWa1WWa1W+frmfarPx8dHNpvnreJOhc/JrMZHPiw2bmfl8dZ8Lv13IwAAAORhs5zbXBW7GCpUqKCbbrpJI0eO1EsvvaTQ0FC99dZbOnjwoDp27OiaPl4GMhMAAAAAKIbFixcrJiZG3bp1U5MmTfTVV1/p448/1jXXXOPuruVDhQ8AAACAZ/OwdfiqVq2q9957z/l9cQESPifLsfnI10bh9LwcHx7pBAAAANyFhA8AAACARzO2c5urYnszSlEAAAAA4KWo8DkZC6/nlWty3N0FAAAAlHS2vzZXxfZiVPgAAAAAwEtR4QMAAADg2Yzl3Oaq2F6MhM/JcoxFPl7+oSkOK4vQAwAAAG5DwgcAAADAszGGz2GUXwAAAADAS1Hhc7KzNh+JhdftcrgXAAAAuFzmr81Vsb0YCR8AAAAAz2aznNtcFduLUX4BAAAAAC9FhQ8AAACARzPm3Oaq2N7M6yp8I0aMkMVi0bx58/LsT01NVXx8vIKCglS5cmVNnTo133sXLFigmJgYBQYGqk2bNtqzZ0+xz59t82W7YLMaH7a/bQAAAMCV4lXfPhMTE7Vq1SpVrlw5X1uvXr30+++/KykpSXPmzNG0adM0f/78PO8dPHiwxo4dqw0bNigqKkpdu3bV2bNnr+QlAAAAAPi782P4XLV5Ma9J+E6cOKFBgwZpwYIF8vf3z9O2bds2rVmzRvPmzVOTJk3UvXt3Pfroo3rppZfsx8yePVt33XWXBg0apAYNGmj+/Pn69ddf9fnnn1/pSwEAAAAAp/CaMXwPP/yw+vTpo+uvvz5f2/r16xUdHa169erZ98XFxWnKlCnKyspSYGCg1q9fr4kTJ9rby5UrpxYtWig5OVnx8fH5YlqtVuXk5NhfZ2VlSZKycy0yFu/+K0FxnGVZBgAAAFwuYzm3uSq2F/OKb+NLly7V9u3bNX78+ALb09LSFBERkWdfeHi4bDabjh07dtFj0tLSCow5ZcoUBQUF2bfQ0FAnXAkAAAAAOI9HJ3xDhw6VxWIpdGvbtq3S09P18MMP6+2335afn1+BcYwLpt4ZN26cMjMz7VtGRobTzwEAAABAMjbXbt7Mox/pnD59eqFVO0kKCAjQjh07dOTIkTyPcubm5mrIkCF666239N133ykyMjJfpS49PV0+Pj4KCwuTJEVERBR4TO3atQs8t5+fX4EJZrbxkeExRjse6QQAAADcx6MTvpCQEIWEhFz0mBtuuEHbt2/Ps69Tp04aMmSI+vTpI0mKjY3V4cOHtXfvXtWtW1fSuVk5GzZsqMDAQPsxK1eu1MCBAyVJmZmZSk5O1ogRI5x8VQAAAACKhTF8DvPohK8oypUrpwYNGuTZ5+fnpypVqigmJkaS1KhRI7Vu3VqDBg3SrFmzlJKSohkzZmjmzJn29wwfPlydO3dWu3bt1LJlS02ePFlVqlRRly5druTlAAAAAIDTlPiEr6gWL16sIUOGqFWrVqpQoYJGjx6tAQMG2Nvj4uI0d+5cTZo0SampqWrRooUSEhLyLfFwKWdzLTKePTTyisphoXEAAABcLip8DvPKhC8lJSXfvqioKC1btuyi7xswYECeJBAAAAAASjKvTPgAAAAAeA9XzqbJLJ0olrM2Fl6/0Fkb9wIAAABwFxI+AAAAAJ6NMXwOY0YNAAAAAPBSVPicLDvXRzbyaLscHukEAADAZTLGIuOi75WGCh8AAAAAoCSiwgcAAADAszGGz2EkfE52buF17/7QFMdZG0VkAAAAwF1I+AAAAAB4NGMsLhtrxxg+AAAAAECJRIXPyc4yS2ce2czSWQDj7g4AAACULDbLuc1Vsb0YmQkAAAAAeCkqfAAAAAA8GmP4HEfC52TZub7Kla+7u+ExzuRQRM4v190dAAAAQClBwgcAAADAs7EOn8MovwAAAACAl6LCBwAAAMCjMYbPcSR8Tpad46NcQ+H0vDO53Iv8GMMHAACAK4OEDwAAAIBHMzaLjIvWy3NVXE9B+QUAAAAAvBQVPic7a/ORzUIefd6ZHJaoyM/q7g4AAACULMzS6TAyEwAAAADwUlT4AAAAAHg0Zul0HAmfk51lls48snK4FwAAAIC7kPABAAAA8GhU+BxH+QUAAAAAvBQVPifLsvnIlzzajlk6AQAAcLmo8DmOzAQAAAAAvBQVPgAAAAAezRgfGRdNjGiMcUlcT0HC52RZNomHGP8ni0c6AQAAALch4QMAAADg0RjD5zjG8AEAAACAl6LC52RnjE0+xububngMHukEAADAZbNZzm2uiu3FqPABAAAAgJeiwgcAAADAozGGz3EkfE52WmfkIx7pPO90bjl3dwEAAAAotUj4AAAAAHg0Y1xXifPyZfgYwwcAAAAA3ooKn5NZLTnysZBHn2eTl//JBAAAAC5n5CPjolqVq+J6Cu++OgAAAAAoxajwAQAAAPBozNLpOBI+J8tRjnzk3R8aAAAAACUDCR8AAAAAj2YzFtlcVIlzVVxPwRg+AAAAAPBSVPgAAAAAeDTG8DmOhM/JcmSlbHoBm7s7AAAAAJRiJHwAAAAAPJoxPjLGRevwuSiup/DuqwMAAACAUowKn5PZZJVxdycAAAAAL8IYPsdR4QMAAAAAL0WFDwAAAIBHo8LnOK9J+Pbv369Ro0YpMTFRxhg1bdpUiYmJKlPm3CWmpqZq6NCh+vrrr1WxYkU9/PDDevzxx/PEWLBggSZPnqyjR48qNjZWb7zxhurVq1esfuSas7IYHuo8b/bEDHd3AQAAACi1vCLhS09P180336wePXpo9erVCg4O1pYtW2Sx/C9b79Wrl4wxSkpK0sGDB3X//fcrKipKAwYMkCQlJiZq8ODBmjNnjlq1aqXJkyera9eu2rFjh/z9/d11aQAAAECpR4XPcV6R8E2fPl1XX321XnnlFfu+unXr2v9727ZtWrNmjXbv3q169eqpSZMmevTRR/XSSy/ZE77Zs2frrrvu0qBBgyRJ8+fPV3h4uD7//HPFx8df0esBAAAA8D9GPrK5aPoR4+XTmnjF1SUkJKhJkybq1q2bIiIidNNNN2n16tX29vXr1ys6OjrP45lxcXHavn27srKy7Me0b9/e3l6uXDm1aNFCycnJBZ7TarUqKysrzyZJubKyXbABAAAAcB+vSPhSUlL06quvqmXLlvrqq6/Upk0bderUSQcPHpQkpaWlKSIiIs97wsPDZbPZdOzYsYsek5aWVuA5p0yZoqCgIPsWGhrqgisDAAAAYGSxP9bp9E3e/UinRyd8Q4cOlcViKXRr27atJMlms6lVq1Z6/PHH1bRpU02dOlXXXHON3n33XUmSccEkKuPGjVNmZqZ9y8hgchIAAAAAnsWjx/BNnz5d48ePL7Q9ICBAkhQZGan69evnaatfv74OHTpkb/97pS49PV0+Pj4KCwuTJEVERBR4TO3atQs8t5+fn/z8/PLtz5XVy/9GAAAAAFxZnjhpy6ZNm/Tvf/9b69atU0BAgG699VYtWbLEyb27fB5d4QsJCVF0dHShW3h4uCSpZcuW2rdvX5737tu3T9WrV5ckxcbG6vDhw9q7d6+9PTExUQ0bNlRgYKD9mJUrV9rbMzMzlZycrBYtWrj6MgEAAACUID/99JPat2+vm2++WRs2bFBSUpJ69+7t7m4VyKMrfEU1YsQItWnTRi+99JK6dOmiJUuWaOfOnfroo48kSY0aNVLr1q01aNAgzZo1SykpKZoxY4ZmzpxpjzF8+HB17txZ7dq1U8uWLTV58mRVqVJFXbp0cdNVAQAAAJD+GsPnoufoHIk7fvx4de/eXRMnTrTvu+aaa5zZLafx6ApfUd18881auHChZs+erUaNGumTTz7Rl19+qRo1atiPWbx4sSpWrKhWrVppyJAhGj16tH1JBuncrJ1z587V5MmT1axZMx09elQJCQnFXoMv11iVa86y/bUBAAAAJcHfZ+C3WguecT43N1dffvmlatWqpbZt2yoyMlK33nqrtm3bdoV7XDQW44oZTUqhrKwsBQUFqWKTobL4eEXh1CnWTKrp7i4AAADgAmeyrYr951hlZmbahzd5qvPfsX8cPkBly7jmO/aZnBw1eGV+vv0TJkzQU089lW9/amqqKleurODgYD3//PO64YYbNHv2bH366afat2+fKlas6JJ+OorMBAAAAECpl5GRkScBLlNIgmmz2SRJPXv21JAhQyRJc+fO1Weffably5frvvvuc31ni4GEz8lsxioLRVMAAADAaYzxkTGuGY12Pm5gYGCRKp5hYWHy9fXNs0qAn5+fYmJi7KsEeBKvGMMHAAAAAFeCv7+/mjZtmmeVgJycHKWkpNhXCfAkVPgAAAAAeDRPm6Xz0Ucf1cCBA9WuXTvdcMMNeumllyRJ3bp1c3b3LhsJn5PZTLYsJtfd3QAAAADgIvfcc4/S09M1duxY/fHHH2revLm++eYbVahQwd1dy4eEDwAAAIBHsxmLbMY1FT5H444YMUIjRoxwal+OHj2qpKQk/fzzz8rKylJYWJgaN26s5s2bFzqJzKWQ8AEAAACAGy1atEhz5sxRUlKSoqKiVLlyZQUGBur333/XgQMHVK5cOd1zzz0aNWqUatasWazYJHwAAAAAPJoxFhkXVfhcFbeoGjZsqNDQUPXt21dLlixRlSpV8rSfPXtW69ev14cffqhWrVrpxRdf1N13313k+CR8TmaUK7loQCkAAAAA7zJ37lzdeOONhbb7+/vr5ptv1s0336ynn35aP//8c7Hik/ABAAAA8GieNkunM10s2fu74OBgXXfddcWKzzp8AAAAAOBGGzduVFxcnE6ePJmv7eTJk4qLi9PmzZsdik3CBwAAAMCjnZ+l01Wbuz377LPq2LFjgcs6VKhQQZ07d9a0adMcik3CBwAAAABulJycrH/84x+Ftnft2lU//PCDQ7EZwwcAAADAo3nzLJ2SlJaWpvLlyxfa7ufnp/T0dIdik/A5mTE2ydjc3Q0AAAAAJUT16tW1efNmVatWrcD2TZs2qXr16g7F5pFOAAAAAB7NJotLN3e78847NW7cOB07dixf27FjxzRu3DjdeeedDsWmwgcAAAAAbvT444/r66+/Vu3atdWnTx/Vr19fkrR7924tXLhQdevW1eOPP+5QbBI+AAAAAB7N28fwBQUFac2aNXr++ef1wQcfaMGCBZKkOnXq6P/+7/80atQoBQYGOhSbhA8AAAAA3Kxs2bIaP368xo8f79S4JHwAAAAAPJorx9p5whg+V2LSFiczsrFdsAEAAAC4uKSkJF133XWKiYnRf/7zH6fGpsIHAAAAwKMZ47qxdsa4JGyxPPjgg5o2bZoaN26sa665Rj179nR4zN7fUeEDAAAAADc6ffq0QkJCVLFiRVmtVlmtVqfFpsLnbCZX8oCZfgAAAABvYWSRcdFYO1fFLY7nnntOffr0UZkyZfT444+rQoUKTotNwgcAAAAAbtS9e3d17dpV2dnZKl++vFNjk/ABAAAA8Gg2Y5HNRU/RuSpucfn7+8vf39/pcRnDBwAAAABusmjRIpkizhyTkpKi7777rljxSfgAAAAAeLTzY/hctbnTe++9pzp16uiJJ57QunXrdObMmTztP//8sxYtWqT4+HjddNNNOn36dLHiO/xI5++//66srCyFhoaqbNmyjoYBAAAAgFLr008/1Q8//KBXX31VnTp1UmZmpipUqKCAgACdOHFC2dnZatiwofr27at3331XwcHBxYpf5ITv9OnTev/99/XBBx9o3bp1OnXqlL2tbt266tChgwYMGKDrr7++WB3wNkZGYsFxAAAAwGm8fQxfy5Yt1bJlSy1YsEBbt27Vzz//rDNnzig0NFSNGzdWRESEw7GLlPA9++yzevbZZ3X11VerS5cuGjlypCpXrqzAwED9/vvv2rlzp77//nt16tRJTZs21axZs3TNNdc43CkAAAAAKG18fHzUtGlTNW3a1Gkxi5Tw7du3T8nJyapdu3aB7bGxserXr59ee+01vffee9q8eTMJHwAAAAAnceVYO/dX+FypSAnf66+/XqRgvr6+6tOnz2V1qMQzNhZeBwAAAOARLmsdPmNMvilEfXyY+BMAAACA83j7GD5XKnZ2dujQId15550KDw9XmTJl5Ofnl2cDAAAAAHiGYlf47r77bhljNHv2bEVGRspi8e6MGAAAAIB7uXK9PHevw/d3p06d0vLly3XgwAE99NBDCgkJ0U8//aTQ0FCHZussdsK3ZcsWbdq0SfXq1Sv2yQAAAAAABdu+fbs6duyoChUq6MCBA7rnnnsUEhKid999V4cPH9bbb79d7JjFfqSzVatW2rdvX7FPBAAAAACOOD+Gz1WbpxgxYoQeeOAB7d69W2XLlrXvv/3227Vq1SqHYha7wvfWW29p0KBB2r17t6699tp84/bat2/vUEcAAAAAoDTbuHGj5s2bl29/5cqV9dtvvzkUs9gJ37Zt27R+/Xp9+eWX+dosFotyc3Md6oi3MCzLAAAAADhVaRnDV7FiRaWmpiomJibP/k2bNqlq1aoOxSz2I53Dhg3T3XffraNHj8pms+XZSnuyBwAAAACO6tevn0aMGKGdO3fKYrHoxIkTSkhI0COPPKJBgwY5FLPYFb6MjAw98sgjioyMdOiEAAAAAFAcNnNuc1VsTzFx4kRZLBbdcMMNysrKUvPmzeXv76/hw4drzJgxDsUsdoWvd+/e+uKLLxw6WemQy5ZnAwAAAHApubm52rJli/7v//5Pv//+u3788UetW7dOaWlpev755x2OW+wKX0hIiJ544gl9+eWXatiwYb5JWyZNmuRwZwAAAADg70rDGD4fHx/deOON2rlzp2JiYnTttdc6JW6xE74NGzaoSZMmOn36tH744Yc8bSzCDgAAAADFZ7FY1LhxYx04cCDfpC2Xo9gJ38qVK512cgAAAAC4FFeul+dJ6/A99thjGjFihMaOHasmTZooKCgoT7sjiWCxEz4AAAAAgPPdeeedkqT7779f0v+eoDTGOLwEXpESvo4dO+rxxx9X27ZtL3pcRkaGXn31VYWEhOihhx4qdmcAAAAA4O9Kwxg+STp48KDTYxYp4Rs8eLCGDRum06dPq3Pnzrr++utVuXJlBQQE6Pjx49q1a5e+//57JSUl6b777tMDDzzg9I6WGCy8DgAAAMABNWrUcHrMIiV8PXv2VM+ePZWYmKgPP/xQc+bM0c8//6wzZ84oNDRUjRs31m233aZ3331XERERTu8kAAAAgNLL9tfmqtieYv78+RdtHzBgQLFjFmsMX/v27dW+fftinwQAAAAAcHGTJ0/O89pqtSo1NVVly5ZVRESEQwlfsRde90Rnz57VqFGjFB0draCgIDVp0kRLly7Nc0xqaqri4+MVFBSkypUra+rUqfniLFiwQDExMQoMDFSbNm20Z8+eK3UJAAAAAAphjMWlm6c4ePBgnu3w4cM6evSoOnTooOnTpzsU0ysSvunTp2vx4sV65513tGPHDt1zzz3q1auXdu3aZT+mV69e+v3335WUlKQ5c+Zo2rRpeUqmiYmJGjx4sMaOHasNGzYoKipKXbt21dmzZ91xSQAAAACg8PBwTZ48WY899phD7/eKhC85OVk9e/ZU+/btVatWLT322GOqUKGCtmzZIknatm2b1qxZo3nz5qlJkybq3r27Hn30Ub300kv2GLNnz9Zdd92lQYMGqUGDBpo/f75+/fVXff755266KgAAAACSZJPFpZunO3bsmE6ePOnQe71iHb5WrVpp0aJFOnTokKKjo/Xxxx/r7NmzuummmyRJ69evV3R0tOrVq2d/T1xcnKZMmaKsrCwFBgZq/fr1mjhxor29XLlyatGihZKTkxUfH5/vnFarVTk5OfbXWVlZrrtAAAAAAF7vySefzPPaGKPU1FQtXbpU//znPx2K6RUJ39ixY5WWlqbq1aurTJkyCgwM1EcffaRq1apJktLS0vLNHhoeHi6bzaZjx46pWrVqhR6TlpZW4DmnTJmSJ0EEAAAA4BrGnNtcFdtTrF27Ns9rHx8f+yOdji5951DC9+OPP2rNmjVKS0uTzZZ3ItNJkyY51JGCDB06VHPnzi20vU2bNlq1apXee+89ff7551q2bJlq1qypzz//XHfffbeSkpJUv359GRf8rzhu3DiNHj3a/jorK0uhoaFOPw8AAACA0mHlypVOj1nshG/GjBkaNWqU6tWrp6ioKFks/3vm9cL/dobp06dr/PjxhbYHBARIksaMGaOpU6eqW7dukqRGjRopMTFRr7/+ul544QVFRkbmq9Slp6fLx8dHYWFhkqSIiIgCj6ldu3aB5/bz85Ofn1++/UY2qQQ8BwwAAACUFK4ca+dJY/jat2+vpUuXKiQkJM/+kydPKj4+XomJicWOWeyE7/nnn9fcuXM1aNCgYp+suEJCQvJdbEEyMzPl6+ubZ5+Pj4+9+hgbG6vDhw9r7969qlu3rqRzs3I2bNhQgYGB9mNWrlypgQMH2mMmJydrxIgRTrwiAAAAAMXlyuUTPGlZhlWrVhW4SkBWVpa+//57h2IWO+E7c+aM2rVr59DJXKVLly566qmnVLlyZfsjnStWrNC///1vSecqfq1bt9agQYM0a9YspaSkaMaMGZo5c6Y9xvDhw9W5c2e1a9dOLVu21OTJk1WlShV16dLFTVcFAAAAoDR455137P+9ZMkSVahQwf46NzdXa9asKfTJw0spdsI3bNgwvfnmm5o2bZpDJ3SF2bNna8yYMbrvvvv0xx9/qHbt2lqwYIHi4uLsxyxevFhDhgxRq1atVKFCBY0ePTrPSvVxcXGaO3euJk2apNTUVLVo0UIJCQny9/d3xyUBAAAA+Ivtr81Vsd1t3Lhx9v+eNm2afHz+t3qen5+fatSooVdffdWh2BZThBlN7r///jyvly1bpurVq6tBgwb5xrFdmJ2WJllZWQoKCpLfNR1l8fG99BtKiY3PxF36IAAAAFwxZ7Ktiv3nWGVmZtqHN3mq89+x37/3CQWUyT9/hjNk51jVe+Fkj7gf7dq109KlS3XVVVc5LWaRKnx/Hx/Xo0cPp3UAAAAAAC6mtIzhc9ssnQsWLHD6iQEAAAAAee3YsUMff/yxDh06JKvVmqdt/vz5xY7nc+lD8mrfvr2OHz+eb//JkyfVvn37YncAAAAAAC7G5uLNUyxZskTNmjXT999/r7feekupqan6/vvv9dFHH+VL/oqq2AmfK6YKBQAAAIDSbvLkyXrppZf0xRdfyN/fX7Nnz9auXbt03333qVq1ag7FLPIsna6cKhQAAAAAClNaxvAdOHBAHTt2lCSVLVtWf/75pywWi/71r3/ppptu0tSpU4sds8gJnyunCvUqxiZ50IcGAAAAQMkQFRWljIwM1axZUzVr1tTatWvVuHFj7d27VzabYw+fFjnhO3TokCTXTBUKAAAAAIUxct1Yu0uuUXcFdevWTV9++aWaNWumhx56SIMHD9b8+fO1e/duDRo0yKGYxV54/cKpQv/44w9JIvkDAAAAgMs0Y8YM+3/37dtXMTExWr9+vWrXrq34+HiHYhZ70pacnBxNmjRJERERCgsLU1hYmCIiIjRp0iSHZ44BAAAAgMIYWVy6eYKzZ8/qjjvu0P79++37brnlFo0aNcrhZE9yoMI3bNgwJSQkaNq0aYqNjZUkrV+/Xk899ZQOHz6s119/3eHOAAAAAEBp5O/vr6SkJIfH6hWm2Anfe++9p08++URxcXH2fQ0bNlTNmjUVHx9PwgcAAADAqWzGIpuLJkZ0VVxHPPDAA5ozZ06eRzsvV7ETvquuukqRkZH59oeHh6tixYpO6VSJxiydAAAAABxw6NAhLVu2TJ999pkaN26soKCgPO0XLpVXVMUewzd16lT961//0t69e+379u7dq5EjRzq0LgQAAAAAXIxx8eYp/Pz81LNnT918880qX768fH1982yOKHaFb+zYscrIyNDVV1+t8uXLy2Kx6OTJkwoICNCePXs0fvx4+7G//PKLQ50CAAAAgNJmwYIFTo9Z7ITv6aefdnonAAAAAKAwNnNuc1VsT2KM0bp163TgwAHFx8crODhYf/zxh4KCghQQEFDseMVO+Pr27VvskwAAAAAALu7nn3/WP/7xDx04cEBnzpzRnj17FBwcrAkTJignJ0dz5swpdsxij+GTzj2qOXXqVD3wwANKT0+XJK1atSrPuD4AAAAAcIbSsA6fJD300EOKjY3VH3/8ocDAQPv+nj176uuvv3YoZrETvtWrV+vaa6/V6tWr9Z///Ed//vmnJCk5OVljx451qBMAAAAAUNp99913Gj16tPz8/PLsr169un799VeHYhY74Xvsscf0zDPP6KuvvpK/v799f1xcnNatW+dQJwAAAACgMOfH8Llq8xR+fn46depUvv179uxRWFiYQzGLnfD9+OOP6tq1a779lSpVUkZGhkOdAAAAAIDS7s4779TYsWN14sQJSZLFYtGOHTs0atQo9e7d26GYxU74oqKiChyrt2bNGsXExDjUCW9iZGO7YAMAAAAuV2kZw/f8888rIiJCkZGRyszMVKNGjdSoUSNdffXVmjJlikMxiz1L54gRIzRs2DDNmjVLkrRz50598cUXeuKJJ/Tss8861AkAAAAAKO0CAwP1zjvvaNKkSdq5c6dOnTqlxo0bq379+g7HLHbC969//UvBwcF6+OGHdfr0aXXr1k1RUVGaNGmSHnjgAYc7AgAAAAAFKU3r8ElSzZo1VbFiRUnSVVdddVmxHFqWYcCAAdq/f7/+/PNPpaam6siRI3rooYcuqyMAAAAAUJrl5ORo0qRJioiIUFhYmMLCwhQREaFJkybJarU6FLPYFb7c3Fxt2rRJKSkpslgsqlWrlsLCwuTj41DuCAAAAAAX5cqxdp40hm/YsGFKSEjQtGnTFBsbK0lav369nnrqKR0+fFivv/56sWMWK+FLSEjQgw8+qMOHD+fZX716dc2dO1edOnUqdgcAAAAAoCSKj4/XsmXLtGLFCnXo0OGy47333nv65JNPFBcXZ9/XsGFD1axZU/Hx8Q4lfEUuy23btk09evRQp06dtGXLFp05c0ZZWVnatGmT4uLiFB8frx9//LHYHQAAAACAi/HEdfgWLFigrKwsp17nVVddpcjIyHz7w8PD7WP6iqvICd+MGTN055136o033lCjRo3k7++vgIAANWnSRG+++aZ69OihF1980aFOAAAAAEBJ8fPPP2vChAl68803nRp36tSp+te//pVnGby9e/dq5MiRmjp1qkMxi/xI59q1azV//vxC2wcPHqyBAwc61AkAAAAAKIz5a3NVbEn5qnVlypSRn59fvuNtNpv69u2riRMnKjo62ql9GTt2rDIyMnT11VerfPnyslgsOnnypAICArRnzx6NHz/efuwvv/xSpJhFTviOHDly0YXVY2JidOTIkaKGAwAAAACPERoamuf1hAkT9NRTT+U7bsaMGQoODlb//v2d3oenn37a6TGLnPCdOXNG/v7+hbb7+/srOzvbKZ0q0YxNMp4z0w8AAABQ0tmMRTYXfcc+HzcjI0OBgYH2/WXK5E+VfvrpJ73wwgvauHGjS/rSt29fp8cs1iydzzzzjMqVK1dg2+nTp53SIQAAAAC40gIDA/MkfAVJTk5Wamqqqlevnmd/p06d1Lt3by1cuPCy+5GTk6Pdu3crPT1dNpstT1v79u2LHa/ICV/r1q21adOmSx4DAAAAAM50JcbwFUV8fLyaN2+eZ1/Dhg01d+5cde7c+bL78s033+j+++9XampqvjaLxaLc3Nxixyxywrdq1apiBwcAAAAAbxESEqKQkJB8+2vWrOmUCVyGDRumHj166IknnihweQZHFOuRTgAAAAC40ow5t7kqtqf47bff9Oijjzot2ZOKsQ4fAAAAACAvY4w6dOjglFj33HOPvvzyS6fEOo8KHwAAAACPZpNFNrlolk4XxXXErFmz1K1bN3311Ve67rrr8q0DOGnSpGLHJOEDAAAAAA/w4osv6uuvv1b9+vV18uRJWSz/S0Yv/O/iIOEDAAAA4NGMXDiGzzVhHfLMM8/orbfe0v333++0mIzhAwAAAAAPEBgYqJYtWzo1JgkfAAAAAI9mc/HmKcaMGaNnnnlGVqvVaTF5pBMAAAAAPMAHH3ygbdu2admyZapbt26+SVvWrFlT7JgkfM5mbJLxnJl+AAAAgJLOGIuMi75juyquIzp06OC0JR7OI+EDAAAAAA8wYcIEp8dkDB8AAAAAj2ZcvHmSU6dOadGiRXr66ad1/PhxSdJPP/2ktLQ0h+JR4QMAAAAAD7B9+3bdeuutqlixog4cOKB77rlHISEhevfdd3X48GG9/fbbxY5JhQ8AAACARzNGsrloc9X6fo4YMWKEBg0apN27d6ts2bL2/bfffrtWrVrlUEwSPgAAAADwABs3blT//v3z7a9cubJ+++03h2KS8AEAAADwaEYWl26eomLFikpNTc23f9OmTapatapDMUn4AAAAAMCN1qxZI6vVqn79+mnEiBHauXOnLBaLTpw4oYSEBD3yyCMaNGiQQ7GZtAUAAACARzs/3s5Vsd2tXbt2Onr0qCZOnCiLxaIbbrhBWVlZat68ufz9/TV8+HCNGTPGodgkfAAAAADgRuavmWN8fHw0adIkjRs3Tvv379epU6d0zTXXqHz58g7H9vhHOtesWaMuXbooPDxcFotF+/bty3dMamqq4uPjFRQUpMqVK2vq1Kn5jlmwYIFiYmIUGBioNm3aaM+ePcWOAQAAAODKKw3r8Fks/xtLGBAQoGuvvVaxsbGXlexJJaDCd/r0aTVv3lzdu3fX4MGDCzymV69eMsYoKSlJBw8e1P3336+oqCgNGDBAkpSYmKjBgwdrzpw5atWqlSZPnqyuXbtqx44d8vf3L1IMAAAAAHCV7t2723OTwiQmJhY7rscnfLfddptuu+02paSkFNi+bds2rVmzRrt371a9evXUpEkTPfroo3rppZfsydrs2bN111132Qc6zp8/X+Hh4fr8888VHx9fpBgAAAAA3MMYyRjXzKbpKevwxcbGqly5ck6P6/EJ36WsX79e0dHRqlevnn1fXFycpkyZoqysLAUGBmr9+vWaOHGivb1cuXJq0aKFkpOTFR8fX6QYf2e1WpWTk2N/nZWV5aIrBAAAAODtxowZo4iICKfH9fgxfJeSlpaW78aEh4fLZrPp2LFjFz0mLS2tyDH+bsqUKQoKCrJvoaGhzrokAAAAABewuXhztwvH7zmb2xK+oUOHymKxFLq1bdu2SHGME2qwjsQYN26cMjMz7VtGRsZl9wMAAABA6eOMnKYwbnukc/r06Ro/fnyh7QEBAUWKExkZaa/UnZeeni4fHx+FhYVJkiIiIgo8pnbt2kWO8Xd+fn7y8/MroMUmyXUZOgAAAFDanBvD57rY7mazua7O6LYKX0hIiKKjowvdwsPDixQnNjZWhw8f1t69e+37EhMT1bBhQ/vYu9jYWK1cudLenpmZqeTkZLVo0aLIMQAAAACgpPH4SVtOnTqlffv26ciRI5Kkn376SadOnVL16tVVqVIlNWrUSK1bt9agQYM0a9YspaSkaMaMGZo5c6Y9xvDhw9W5c2e1a9dOLVu21OTJk1WlShV16dJFkooUAwAAAIB7GFlkXPQUnaviegqPn7Rl48aNatq0qbp27SpJ6tatm5o2barly5fbj1m8eLEqVqyoVq1aaciQIRo9enSe5RTi4uI0d+5cTZ48Wc2aNdPRo0eVkJCQZ52LS8UAAAAAgJLG4yt8bdu2veQgxqioKC1btuyixwwYMOCiCVxRYgAAAAC48mzm3Oaq2N7M4yt8AAAAAADHeHyFDwAAAEDp5u2zdLoSCR8AAAAAj2aTRTYXTa7iqriegkc6AQAAAMBLUeEDAAAA4NF4pNNxVPgAAAAAwEtR4QMAAADg0cxfm6tiezMSPmczNsl498BPAAAAACUDCR8AAAAAj2YzFtlcVFRxVVxPwRg+AAAAAPBSVPgAAAAAeDRm6XQcFT4AAAAA8FJU+AAAAAB4NGbpdBwVPgAAAADwUlT4AAAAAHg0Zul0HBU+AAAAAPBSVPgAAAAAeDTG8DmOCh8AAAAAeCkqfAAAAAA8GuvwOY4KHwAAAAB4KSp8AAAAADwas3Q6jgofAAAAAHgpKnwAAAAAPJ6XD7VzGSp8AAAAAOClqPABAAAA8GjGWGRcNNbOVXE9BRU+AAAAAPBSVPiczBib5OV/JQAAAACuJNtfm6tiezMqfAAAAADgpajwAQAAAPBoxpzbXBXbm1HhAwAAAAAvRYUPAAAAgEdjlk7HUeEDAAAAAC9FhQ8AAACAR2OWTsdR4QMAAAAAL0WFDwAAAIBHYwyf46jwAQAAAICXosIHAAAAwKPZzLnNVbG9GRU+AAAAAPBSVPgAAAAAeDQji4xcNIbPRXE9BRU+AAAAAPBSVPgAAAAAeDRjzm2uiu3NqPABAAAAgJeiwgcAAADAo9mMRTYXrZfnqrieggofAAAAAHgpKnxOZyTZ3N0JAAAAwGuYvzZXxfZmVPgAAAAAwEtR4QMAAADg0RjD5zgqfAAAAADgpajwAQAAAPBorMPnOCp8AAAAAOClqPABAAAA8GhGFhm5Zqydq+J6Co+v8K1Zs0ZdunRReHi4LBaL9u3bl6c9JSVF/fv3V40aNRQYGKhrrrlGr776ar44CxYsUExMjAIDA9WmTRvt2bMnT3tqaqri4+MVFBSkypUra+rUqS69LgAAAABwNY9P+E6fPq3mzZsXmoDt2rVLvr6+mj9/vnbs2KHx48dr1KhReuedd+zHJCYmavDgwRo7dqw2bNigqKgode3aVWfPnrUf06tXL/3+++9KSkrSnDlzNG3aNM2fP9/l1wcAAADg4mzGtZs3sxhTMoYppqSkqFatWtq7d6/q1Klz0WOHDBmi9PR0LV26VJLUo0cPBQYGauHChZLOJZHh4eFatGiR4uPjtW3bNjVu3Fi7d+9WvXr1JElPPvmkli9fri1bthSpf1lZWQoKCpJv7Rtk8fH4PPqK2TTjTnd3AQAAABc4k21V7D/HKjMzU4GBge7uzkWd/47d+6bZKuPr75Jz5OSe1fvfP1Qi7ocjvDIzOXbsmCpVqmR/vX79erVv397+uly5cmrRooWSk5Pt7dHR0fZkT5Li4uK0fft2ZWVlFXgOq9WqrKysPBsAAAAA5zPG4tKtOKZOnarrr79ewcHBqly5svr376/09HQXXfnl87qELzk5WZ999pkGDBhg35eWlqaIiIg8x4WHhystLe2i7TabTceOHSvwPFOmTFFQUJB9Cw0NdfKVAAAAAPA03333nUaOHKmNGzdq2bJl2rlzp3r16uXubhXKbQnf0KFDZbFYCt3atm1b7Jh79uzRHXfcoYkTJ+rGG28s8vsceap13LhxyszMtG8ZGRnFjgEAAADg0mwu3orj888/V58+fXT11VcrNjZWM2fO1MqVK3XixInLu0gXcduyDNOnT9f48eMLbQ8ICChWvAMHDiguLk4DBgzQmDFj8rRFRETYq3nnpaenq3bt2pKkyMjIAtt9fHwUFhZW4Pn8/Pzk5+dXrD4CAAAA8Ex/H6JVpkyZIn3fP3bsmMqWLaty5cq5qmuXxW0VvpCQEEVHRxe6hYeHFznWL7/8ovbt2ys+Pr7A2TxjY2O1cuVK++vMzEwlJyerRYsW9vbDhw9r79699mMSExPVsGFDrxy4CQAAAJQkV2IMX2hoaJ4hW1OmTLlkv7KzszVp0iT17dtXZcp45hLnntmrC5w6dUr79u3TkSNHJEk//fSTTp06perVq6tSpUr69ddf1a5dOzVu3FiPP/64UlNTJUn+/v72iVuGDx+uzp07q127dmrZsqUmT56sKlWqqEuXLpKkRo0aqXXr1ho0aJBmzZqllJQUzZgxQzNnznTLNQMAAAC4sjIyMvIUey6VwOXm5qpPnz6SpOeff96lfbscHp/wbdy4Ue3atbO/7tatm6RzC6n369dPK1as0IEDB3TgwAEtX77cflybNm20atUqSedm3Jw7d64mTZqk1NRUtWjRQgkJCfL3/9/UrosXL9aQIUPUqlUrVahQQaNHj84z8QsAAAAA93Dlennn4wYGBhb56T6bzaZ+/fpp165dWr16tYKDg13TOScoMevweTrW4SsY6/ABAAB4lpK4Dl+PlnNcug7f0h+GFfl+GGM0cOBArV27VmvXrlVUVJRL+uUsHl/hAwAAAFDaWWRUvPXyihO7OIYOHapPP/1UCQkJkmQfUhYeHi5fX1+n9+5ykfA5m7FJ1EwBAAAAr/T6669Lkn0CyPMOHjyomjVruqFHF0fCBwAAAMCj2YxkcfEYvqIqaSPiGGwGAAAAAF6KCh8AAAAAj2bMuc1Vsb0ZFT4AAAAA8FJU+AAAAAB4NJsssrholk6by2b/9AwkfAAAAAA8mk0unLTFNWE9Bo90AgAAAICXosIHAAAAwKMZY5Exrnn00lVxPQUVPgAAAADwUlT4AAAAAHg0m+SyqVUYwwcAAAAAKJGo8AEAAADwaDbjwlk6WXgdAAAAAFASUeEDAAAA4NFYeN1xVPgAAAAAwEtR4QMAAADg0Yw5t7kqtjejwgcAAAAAXooKHwAAAACPxjp8jqPCBwAAAABeigofAAAAAI/GOnyOo8IHAAAAAF6KCh8AAAAAj2aTkUWuKcXZXBTXU1DhAwAAAAAvRYUPAAAAgEdjlk7HUeEDAAAAAC9FhQ8AAACAR2OWTsdR4QMAAAAAL0WFDwAAAIBHy5WRXDSbZi6zdAIAAAAASiIqfAAAAAA8mpFx2Xp5xssrfCR8zmZsrqo2AwAAAECxkPABAAAA8Gg2F47hc1Xl0FMwhg8AAAAAvBQVPgAAAAAejQqf46jwAQAAAICXosIHAAAAwKPlyiYjm0ti21wU11NQ4QMAAAAAL0WFDwAAAIBHO1eFo8LnCCp8AAAAAOClqPABAAAA8GhU+BxHhQ8AAAAAvBQVPgAAAAAejQqf46jwAQAAAICXosIHAAAAwKPlWmwyFhdV+FwU11NQ4QMAAAAAL0WFDwAAAIBHYwyf46jwAQAAAICXosIHAAAAwKOdq8LlujC296LCBwAAAABeyuMTvjVr1qhLly4KDw+XxWLRvn37Cj32l19+UcWKFRUdHZ2vbcGCBYqJiVFgYKDatGmjPXv25GlPTU1VfHy8goKCVLlyZU2dOtXp1wIAAACg+Gwu/j9v5vEJ3+nTp9W8efNLJmDGGPXt21etWrXK15aYmKjBgwdr7Nix2rBhg6KiotS1a1edPXvWfkyvXr30+++/KykpSXPmzNG0adM0f/58p18PAAAAAFwpHp/w3XbbbZo0aZJuvfXWix43Y8YMVapUSb17987XNnv2bN11110aNGiQGjRooPnz5+vXX3/V559/Lknatm2b1qxZo3nz5qlJkybq3r27Hn30Ub300ksuuSYAAAAARWdTrks3b+bxCV9R7Ny5UzNnztSrr75aYPv69evVvn17++ty5cqpRYsWSk5OtrdHR0erXr169mPi4uK0fft2ZWVlFRjTarUqKysrzwYAAAAAnqTEJ3xWq1X33XefXnjhBUVERBR4TFpaWr628PBwpaWlXbTdZrPp2LFjBcacMmWKgoKC7FtoaKgTrgYAAADA3xnlunTzZm5L+IYOHSqLxVLo1rZt2yLFefrpp1W3bl3deeedDvfFGFPs94wbN06ZmZn2LSMjw+HzAwAAAIAruG0dvunTp2v8+PGFtgcEBBQpzurVq7V27Vp9+OGHks4lbzabTWXKlNHnn3+ujh07KiIiwl7NOy89PV21a9eWJEVGRhbY7uPjo7CwsALP6+fnJz8/vyL1EQAAAIDjjAtn0zRePkun2xK+kJAQhYSEXHacBQsW6PTp0/bXy5Yt08svv6xvvvlGNWvWlCTFxsZq5cqVGjhwoCQpMzNTycnJGjFihL398OHD2rt3r+rWrSvp3MyeDRs2VGBg4GX3EQAAAADcwW0JX1GdOnVK+/bt05EjRyRJP/30k06dOqXq1aurUqVKqlWrVp7jN27cqDJlyqhBgwb2fcOHD1fnzp3Vrl07tWzZUpMnT1aVKlXUpUsXSVKjRo3UunVrDRo0SLNmzVJKSopmzJihmTNnXrHrBAAAAFAwm3JlcdFoNMbwudnGjRvVtGlTde3aVZLUrVs3NW3aVMuXLy9yjLi4OM2dO1eTJ09Ws2bNdPToUSUkJMjf399+zOLFi1WxYkW1atVKQ4YM0ejRozVgwACnXw8AAAAAXCkW48iMJcgnKytLQUFB8q3VVBYfj8+jr5hNs+52dxcAAABwgTPZVsX+c6wyMzM9fvjS+e/YUU1Hy+LjmvkzjM2q1M3PlIj74QgyEwAAAADwUh4/hg8AAABA6WaTTRZm6XQIFT4AAAAA8FJU+AAAAAB4tHMzaTJLpyOo8AEAAACAl6LCBwAAAMCjsQ6f46jwAQAAAICXosIHAAAAwKOdm0mTWTodQYUPAAAAALwUFT4AAAAAHs0Ym2RcM9bOGCp8AAAAAIASiAofAAAAAI92bpZOi0tie/ssnSR8AAAAADwak7Y4jkc6AQAAAMBLUeEDAAAA4NGMyZWMix7pdNFkMJ6CCh8AAAAAeCkqfAAAAAA8GmP4HEeFDwAAAAC8FBU+AAAAAB6NMXyOo8IHAAAAAF6KCh8AAAAAD+e6MXyui+sZqPABAAAAgJci4QMAAADg0YxyXbo5Yvr06apSpYqCgoLUrVs3paamOvmqnYOEDwAAAACKYcGCBXr66ac1e/ZsJSUl6eTJk+rVq5e7u1UgxvABAAAA8GjG2CTjonX4HIj78ssva8SIEerRo4ckaf78+apdu7a2bNmiJk2aOLmHl4cKHwAAAIBSLysrK89mtVoLPC47O1tbt25V+/bt7ftiYmJUs2ZNJScnX6nuFhkJHwAAAACPZmRz6SZJoaGhCgoKsm9TpkwpsC8ZGRmy2WyKiIjIsz88PFxpaWkuvxfFxSOdAAAAAEq9jIwMBQYG2l+XKVNwqmSMuVJdcgoSPgAAAACezeRKxuK62JICAwPzJHyFCQsLk4+PT75qXnp6er6qnyfgkU4AAAAAKKKAgAA1btxYK1eutO87ePCgUlJS1KJFCzf2rGBU+AAAAAB4NCMjyUWzdKr4j2g+9NBDGjFihJo1a6aYmBg9+uijuuWWWzxuhk6JhA8AAMBjGflIvv7u7gZKotyzsrgoQYI0YMAA/fbbbxo2bJiOHz+uDh066I033nB3twpEwgcAAOBhjCRbULRUvposFh/J4qKxS/BOxpxbW+7PQ/LJPCyv+PQYmwvH8DmWGI8dO1Zjx451cmecj4QPAADAw9iCouUTEqOI8FAF+PvJQsKHYjDGKPusVWm+frJJ8s087O4uwY1I+AAAADyIkY9UvpoiwkNVITjI3d1BCRXg7ydJSs21ymQeKfGPdxoXVviMgxW+koJZOgEAADyJr78sFh/7F3bAUeeqw4wDLe2o8AEAAHgai4XHOHHZLBaLF43/zC2hsd2PCh8AAAAAeCkqfAAAACVETnambLlnXX4eH19/lQlg/CA8iAfO0llSkPABAACUADnZmfpl03IZm+sfP7P4+Kr69d08NunLyclRxfBq+uLTj9T65hvd3Z0i6Xx7D7VqGasJ48c4LWbiqjX6R/deOv3HUafFhPfhkU4AAIASwJZ79ooke5JkbLnFqiR2vr2HJj493YU9ckxmZqbCqtTS0aOp7u4KLpORzaWbNyPhAwAAgFdatfo71atbR5UrR7m7K4DbkPABAADAZXbv2avud96r6rWvVZUa9dX9znuV8vMv9vY13yWp3FWVtXL1WjVr2VqR1eqo17399Mfx4/ZjTpw4qXvuH6jQyrXUuPlN+iZxdZHO/cVXK9S5Y4cC2zJ+/119+g1SdK1rFF41Ri1ujlPy+o329sRVa3Rzu072c859Y4G9LTs7Ww8MfVj1rmum8KoxuqltR61a890l+3MmO1uDHvyXwqvG6OqGzfXRx8vztG/euk2db++h0Mq1dE2jG/T0tOeUk5Njb/9xx0+6uV0nVYqqqQ6du+mXXw4V6T54BWNz7ebFSPgAAADgMqdPn1b8HbdrxRfLtOKLZfL391PfgUPzHffs8zM195VZ+mL5h9qxc5eeeX6mve2xx5/UT7t26/NlH+j1ObM0ZfpzRTr3VysSdVunWwtsmzzlWf156pS+SvhYyd8l6vHRI+X/19qHe/bu0z33D9SgAX21cd0qTX/6KU199gV9uHSZJCknJ1d168Tog/ff1g9rv1XX2zqp1739lJZ+7KL9mf/Wf1Q7pqa+X/W1BvTtowGDh2v/gYOSziWg3br3Vsdb47T++0TNnTNLSz78WLNmvyZJys3N1T33D1S16Kr6buVXGjZ0kCZNfbZI9wGlG5O2AAAAwGWub9pE1zdtYn8968VnVfvqxjp06LCqVYu275/81Hg1b9ZUktT3vnu07NMESdLJk3/q/SUf6YP33laL2OaSpCcef0zd77z3oufduv1HZZ/NVrPrmxTYfvjXI2rV4gZdd+3VkqSYWjXtbS/OekX9+/ZR3/vukSTVqllDDw0dpAXvLFTPHneoXLkgjf6/R+3HPz56lD746GOt+DZR9/a+q9A+XXN1fY3590hJ0mP/94i+/iZR8xa8o2mTJ+j1eW+p9S03auSI4ZKk2jG1NG7M/+npac9p1CMP6ZvEVTpy9KhWf/u5rgoJ0bXX1NfmLdv04qzZF70PXoNZOh1GwgcAAACXOXHipJ6aPE3frlyttPR02Wznvlwf/vVInoTvfOIlSZGREUr/q1p28OeflZOTY08GJeX578J89dU36nRrnHx8Cn6grf/99+r+gUP1beJqtW/XRv/s3k316taRJO3Y+ZN27NylNxe8Yz8+JydXlaMi7a9nvvyqFr3/gY4cOaqz1rPKyjqjXw8fuWifml+ft9/NmjXV3r377edM+OJrRUTXtrfn5tpktVpls9m0d99+xdSqpatCQi64D00ueR8AEj4AAAC4zNgnJmr9hv/q2amTVKNGNeXk5KrlLXGyXjA2TZL8/Pzs/22xWGQzRpJk/vr/FkvxqjtffP2NHh42pND2rl066cfNP+iLL7/WVyu+1bMvzNIbr76knj3u0OnTp/XwsMG6v8/ded5Tpsy5r87vLf5Q0597Uc8/M0WNGlyncuWC1LvPgHzXlM9FLuHUqdPq2eMOjX1sZL42Hx8fGWOKfQ+8ybmZNF1z/d4+SycJHwAAAFxm/Yb/qt/996hzp3OTp3yf9EOx3h9Ts6bKlCmjjf/drFvj2kmS/rtpy0XfcywjQ1u3/agO7dte9LjKUZEa0O8+Deh3nx4ZNUYL31uinj3uUMPrrtPefftVO6ZWge/bsHGT2txyk/rcfe7xzVOnTuvw4V8veS1/7/emTVvUvPn1kqSGDa7TytVrCj1n3Tp1tP/AAR0/cUIhFSsWGA8oCAkfAAAALltaerq2bv8xz77atWopplZNfbLsM8W1a6M//jiucRMmFytuhQrldVfP7hozboJCKlaUMUaTLzFZyVcrvlWL2OaqUKF8occ8Pe05NWvaRFdfXU9//PGH1iVvUJvWN0mSHvnXg2rfqZsmPj1dd/XsLmOk/27eoqzMLA1+oJ9q1aqhjz5Zru+TftBVV12lp6c9K1sRxoHt/GmXnn1+prrH/0OfLE9Q8ob/6tXZMyRJQx7op/lv/UfDR4zSkAcGqGzZAG3/caf27d+v0f/3qG6Na6uoyEgN/9cojR/7b+3avVcL3/+gGHeyhGMMn8OYpRMAAKAE8PH1l8XH94qcy+LjKx9f/2K95613FunG1rfm2TZt2appT0+QMdLN7Trr4Uf/rfFj/13s/jw7bZLq1qmtTrf30IDBwzXmgglTCvLV19/qtk4FL8dwXpkyvnr8yYlq1rKNevS6T82bNdGEcWMkSU2bNNanS9/Xd9+v083tOuvW2+7QuwvfV/Xq1SRJD/S/X21b36IevfroHz166cZWLdTwumsveR39+/bRT7v36MY2t+qNN9/Wm3Nnq07tGElSdHRVfZXwsQ7/ekQdbuum1nG3adbsVxUdXVWS5Ovrq0XvvKmUn3/RjW066uVX5mrc6FGXPCdgMecfjMZlycrKUlBQkHxrNZWlkMHBpdGmWXdf+iAAAGBnfMvKRDRXjWpV5e+X92GsnOxM2XLPurwPPr7+KhMQ5PLzuEJOTo6q175Oq75JsE/CUlqdtebo50O/ypK2UZbcM/b9Z7Ktiv3nWGVmZiowMNCNPby089+xy9S92WV/8DC2XOXs/a5E3A9H8EgnAABACXEuCSuZidiV8vsfxzXq0YdLfbIHnOfxpag1a9aoS5cuCg8Pl8Vi0b59+wo87qWXXlLdunUVEBCgmjVr6t13383TvmDBAsXExCgwMFBt2rTRnj178rSnpqYqPj5eQUFBqly5sqZOneqyawIAAIBrRISHadQjD7m7G3A6m4s37+XxFb7Tp0+refPm6t69uwYPHlzgMZMnT9a8efM0Y8YMNW3aVGlpaXnaExMTNXjwYM2ZM0etWrXS5MmT1bVrV+3YsUP+/ueeT+/Vq5eMMUpKStLBgwd1//33KyoqSgMGDHD5NQIAAACAK5SYMXwpKSmqVauW9u7dqzp1/leiP3bsmKKjo/XVV1+pTZs2Bb63R48eCgwM1MKFCyWdSyLDw8O1aNEixcfHa9u2bWrcuLF2796tevXqSZKefPJJLV++XFu2bClS/xjDVzDG8AEAUDzGt6xMeDNVr1ZVAf5+l34DUIjss1b9cuhXWdL/W/LH8NVp6doxfPt+KBH3wxElPjP55ptv7I961qlTRzExMRoxYoQyMzPtx6xfv17t27e3vy5XrpxatGih5ORke3t0dLQ92ZOkuLg4bd++XVlZWQWe12q1KisrK88GAABw2XLPyhibss9a3d0TlHDZZ60yxiZdgYl+4Lk8/pHOS0lJSVFubq5eeuklvfnmm7JarXrwwQeVlZWl119/XZKUlpamiIiIPO8LDw+3P/pZWLvNZtOxY8dUrVq1fOedMmWKJk6c6KKrAgAApZVFNunPQ0rzPVfdC/D3k8XiovXH4JWMMco+a1Vaeob056Fzn6kSzrhwHT7j5evwuS3hGzp0qObOnVtoe5s2bbRq1apLxrHZbLJarXrppZfsj3Q+//zzuvPOO/Xqq6/K1/fSpV9HnmodN26cRo8ebX+dlZWl0NDQYscBAAD4O5/Mw7JJSs21ymLxkUj4UBzGnEti/jwkn8zD7u4N3MxtCd/06dM1fvz4QtsDAgKKFCcyMlKSVL9+ffu++vXry2q16rffflOVKlUUERGRbyKX9PR01a5d2x6joHYfHx+FhYUVeF4/Pz/5+fFcPQAAcD6LJN/MwzKZR6RiLoAOSJIl96xXVPb+x8h1s2mWiClNHOa2hC8kJEQhISGXHadly5aSpH379ikqKsr+3/7+/vZkMDY2VitXrtTAgQMlSZmZmUpOTtaIESPs7YcPH9bevXtVt25dSedm9mzYsKFXDtwEAAAlg0U26YLJNgCguDx+DN+pU6e0b98+HTlyRJL0008/6dSpU6pevboqVaqk6667TrfeeqseeeQRzZ07Vzk5ORo9erQGDhxof5xz+PDh6ty5s9q1a6eWLVtq8uTJqlKlirp06SJJatSokVq3bq1BgwZp1qxZSklJ0YwZMzRz5kx3XTYAAACA84zNdYU4Lx/D5/GzdG7cuFFNmzZV165dJUndunVT06ZNtXz5cvsxixYtUu3atdWmTRt1795dHTp00AsvvGBvj4uL09y5czV58mQ1a9ZMR48eVUJCgn0NPklavHixKlasqFatWmnIkCEaPXo0a/ABAAAAKNFKzDp8no51+ArGOnwAAACepSSuw+fK79jGZlPuwc0l4n44wuMf6SwpzufNxubdJeHiOpPNGkIAAACe5Pz3sxJV9zE21z156eWPdFLhc5Lff/+dZRkAAABQYmRkZKhSpUru7sZFWa1W1axZ0z6fh6tUqVJFKSkpXjkLPwmfk9hsNh0/flxly5Z16+Ko59cDzMjI8MqStCO4J/lxTy6Ne+QapfW+ltbrLorSdm9K2/UWV2m7P+66XmOMzpw5o5CQEPmUgKFIVqtVOTk5Lj1HmTJlvDLZk3ik02l8fHw86i8kgYGBpeIHZXFwT/Ljnlwa98g1Sut9La3XXRSl7d6UtustrtJ2f9xxvUFBQVf0fJeD9a8vj+en9AAAAAAAh5DwAQAAAICXIuHzMmXKlNGECRNUpgxP657HPcmPe3Jp3CPXKK33tbRed1GUtntT2q63uErb/Slt1wv3YNIWAAAAAPBSVPgAAAAAwEuR8AEAAACAlyLhAwAAAAAvRcLnZaZPn64qVaooKChI3bp1U2pqqru75BJLly5VXFycKlasKIvFUuhinBs3bpSfn59uvvnmfG3eeK+mTp2q66+/XsHBwapcubL69++v9PT0PMcsX75cTZs2VVBQkKKjo/XII48oOzu7wHjx8fGyWCz65ptvrkT3r7i/X9/PP/+s9u3bKyIiQmXLllW9evU0c+bMIr+/tNu0aZPi4uIUFBSkq666SnfddZe9bc+ePWrXrp0CAwNVs2ZNzZ8/P897T5w4oQcffFBVq1ZVuXLl9I9//EOHDx++0pdQLDVr1pTFYsm3LVmypEifpYu93xscP35cAwcOVFRUlIKDg3XjjTdqzZo19vZL/SyaPn26rr76agUFBSk0NFTdunXTnj173HEp+Vzqd9ClPu/SpX8H5eTkaMKECapevboCAgJUr149rVixwqXX5UwXu0dnzpzR/fffr6uvvlo+Pj4aP358vvc/9dRT+f5txMfH5znGk+7Rxa53y5Ytuuuuu1SlShWVK1dOTZs21Ycffpjn/UX5efDFF1+oefPmKleunGrVqqXXXnvtil0fSjYSPi+yYMECPf3005o9e7aSkpJ08uRJ9erVy93dconMzEy1b99eY8aMKfSYrKws9e3bV23bts3X5q336rvvvtPIkSO1ceNGLVu2TDt37sxzXfv371fPnj3Vu3dv7dixQ//5z3/00UcfafLkyfliLViwQFlZWVey+1dUQddXpkwZ3XvvvVqxYoV27dqlSZMmafz48Xr33XeL9P7S7KefflL79u118803a8OGDUpKSlLv3r0lSVarVV27dlVYWJg2bNigJ554QkOGDNG3335rf//AgQO1YcMGffzxx9q4caMCAwN1++23Kzc3112XdEkbNmzQ0aNH7dusWbMUGBiozp07F+mzdLH3e4ORI0dqw4YN+uSTT7R161bFxsbq9ttv1x9//FGkn0W1a9fW7NmztWPHDiUmJsrX11ddu3Z14xX9z8V+BxXl816U30FDhgzRxx9/rHnz5mn37t2aN2+eKleu7PJrc5aL3aPc3FwFBwdr9OjRaty4caExYmNj8/wbeeutt/K0e9I9utj1bt68WdHR0Vq8eLG2b9+u/v37q3fv3lq1apX9mEv9PNi8ebPuuOMO9enTR9u2bdPzzz+v0aNH66OPPrpSl4iSzMBrNG3a1Dz++OP21/v37zeSzObNm93XKRdbuXKlkWSsVmu+tocfftiMHDnSTJgwwdx000152krLvUpKSjKSzPHjx40xxixZssRUrFgxzzEjR440nTp1yrMvJSXFVKtWzRw6dMhIMitWrLhSXb4iinN9PXr0MEOHDnX4/aVFjx49TL9+/QpsW7ZsmQkICDAnT56077vvvvvMHXfcYYwxJjMz0/j6+ppVq1bZ20+ePGksFov58ssvXdpvZ+rQoYO55557Cm0v6LNUnPeXNNdee62ZMWOG/fXJkyeNJLNu3boi/yy60LZt24wkk5qa6qIeF19Bv4Mu9Xk35tK/g7Zt22bKlClj9u3b5/JrcLWL/Z42xpg2bdqYcePG5dtf0O/uC3nqPbrU9Z7XsWNH8+ijjxba/vefB2PHjjVt27bNc8yoUaNMy5YtL6/DKBWo8HmJ7Oxsbd26Ve3bt7fvi4mJUc2aNZWcnOzGnrnHt99+qxUrVmjKlCn52krTvTp27JjKli2rcuXKSZKaNWumrKwsffTRRzLG6NChQ/ryyy/VsWNH+3tsNpv69u2riRMnKjo62l1dd5niXN+2bdv0/fff53kk2NvvjyNyc3P15ZdfqlatWmrbtq0iIyN16623atu2bZKk9evX64YbblD58uXt74mLi7P/e7NarcrNzVVgYKC9PSAgQL6+vkpKSrqyF+OgQ4cOKTExUf369SuwvaDPUnHeXxK1atVKy5Yt07Fjx5Sbm6v58+erSpUqatCgQZF+Fl0oKytLb731lurXr6/w8PArfCXFc6nPe1F+ByUkJKh27dpasmSJqlWrpvr162vixIkeXfF2ha1btyoqKkr16tXT8OHD9ccff9jbSvo9OnbsmCpVqlRgW0E/D7Kzs/P8jJSkoKAgbdy4UVar1ZVdhRcg4fMSGRkZstlsioiIyLM/PDxcaWlpbuqVe5w4cUIPPPCAFixYoLJly+ZrLy33Kjs7W5MmTVLfvn3tC7rGxMTo008/1aBBg+Tv76/q1avr5ptv1siRI+3vmzFjhoKDg9W/f393dd2linJ9N954o8qWLasmTZro4Ycf1r333lus95c26enpyszM1HPPPae7775bX3zxhapVq6a4uDidOHFCaWlpBf57Oz++tEKFCoqNjdXEiROVkZGhM2fOaOzYscrJySkxY2v/85//qEqVKoqLi8uz/2KfpaK8vyR7+eWXFRYWpvDwcAUEBGjatGlKSEhQcHBwkX4WSdJnn32m4OBglStXTgkJCfriiy/k4+PZX10u9Xkvyu+glJQUHTx4UF9//bU+/PBDTZ8+Xa+88oqeeeaZK3MRHqBly5Z65513tGLFCr3wwgtavXq17rjjDpm/lo8uyffoo48+0k8//VSsnwcdOnTQN998o88++0w2m03btm3Tm2++qZycHB07duxKdR0llGf/1ESRnf8BCOlf//qXevXqpZYtWxbYXhruVW5urvr06SNJev755+37jxw5ogcffFCjRo3Sf//7Xy1fvlxff/21nn32WUnnxmG98MILev31193Sb1cr6vUtXrxY//3vfzVv3jzNmDHDPkbC2++Po2w2mySpZ8+eGjJkiK6//nrNnTtXFotFy5cvL9K/uf/85z9KT09XeHi4goODdfjwYV1//fUe/+X+vLffflv33Xdfvv4W9lkq6vtLslmzZmnv3r1asWKFNmzYoLvvvlvdunVTRkbGJX8WndeuXTtt2bJFa9as0TXXXKO7777b46sZl/q8F+Xfg81m09mzZ/XWW2+pRYsW6t69u8aNG6c333zTWd30eJ07d1b37t3VsGFD/eMf/9CyZcu0du1a/fe//5VUcu9RUlKS+vfvr3nz5qlWrVoFHlPQz4PbbrtNTz31lHr16iV/f3/deuutuueeeyTJq35uwDXKuLsDcI6wsDD5+Pjkq1Clp6fn+yuit1u9erUOHz5sT3RsNpuMMSpTpox27NihmjVrevW9stls6tevn3bt2qXVq1crODjY3jZnzhzVqFFD48aNkyQ1atRIf/75px5++GE99thjSk5OVmpqqqpXr54nZqdOndS7d28tXLjwil6LsxX1+qpVqyZJuu6663T06FFNmTJF//znP73+/jgqLCxMvr6+ql+/vn2fn5+fYmJidOjQIUVGRmrXrl153nM+uTuvXr16Wr9+vU6cOKGcnByFhoaqcuXKhX4h8iRJSUnas2dPgY9jFvZZKur7S6qsrCw9+eST+uabb9S6dWtJUtOmTZWQkKBFixbpt99+u+jPovPKlSunOnXqqE6dOoqNjdVVV12lL774Qt26dXPLdRXFpT7vRfl9HRkZqYCAANWoUcPeXr9+fY+fudaVateurZCQEB08eFDNmzcvkfdow4YN6tKli5577jl7svZ3F/t58Pjjj2v06NFKTU1VZGSk5s2bp+DgYIWFhbm45yjpSPi8REBAgBo3bqyVK1faHwE4ePCgUlJS1KJFCzf37sr6+uuvdfbsWfvrOXPm6IcfftA777yjWrVqyd/f32vvlTFGDzzwgH744QetXbs23/iAzMxM+fr65tnn4+Nj/4tzfHy8mjdvnqe9YcOGmjt3rlfMHOjI9dlsNvsjsd5+fxzl7++vpk2bat++ffZ9OTk5SklJUfXq1VWhQgW98MILOnXqlP0PEImJiQX+e6tYsaIkae3atUpNTdXtt99+ZS7iMrz99ttq1aqV6tWrd9HjLvwsOfL+ksRqtcpqtRb488Zms13yZ1Fhzv/xzpPFxsZe9PNelN/XLVu2VHZ2tg4fPmwfK7xv3z77HxBKo19++UXHjx9XzZo1JZW8e7R582Z16tRJ48eP15AhQwo97lI/D3x9fVW1alVJ0ocffqguXbrk+7cE5OOWqWLgEm+++aYJDg42S5cuNVu2bDHt2rUzt9xyi7u75RIZGRlm8+bN5o033jCSzMaNG83mzZvNn3/+me/Ygmb68tZ7NXjwYBMWFmaSk5PN0aNH7VtOTo4xxpgVK1YYHx8fM3PmTLN//36TmJho6tWrZ+67775CY8rLZ6G88Po+/vhjs3DhQvPTTz+ZvXv3mrfffttUrFjRvPjii0V6f2m2cOFCU7ZsWfPuu++a3bt3m+HDh5vIyEhz4sQJk52dbWrXrm3uvPNO8+OPP5o333zT+Pn5mW+++cb+/oSEBLNixQqzf/9+88EHH5iIiAgzbNgwN15R0WRlZZmQkBDz2muv5dlf1M9SYe/3BjfddJOJjY01P/zwg9m7d68ZN26c8ff3Nzt37izSz6LHHnvMJCUlmZSUFJOcnGy6d+9uqlWrZp912J0u9juoKJ/3S/0Oslqt5pprrjGdO3c2P/74o1mxYoWpUqWKeeaZZ9xxuQ651O/pHTt2mM2bN5tmzZqZBx54wGzevNns3bvX/v5///vfZu3atebgwYMmMTHRNGvWzLRq1crk5uYaYzzvHl3serdv325CQ0PNsGHD8vxu/vtn+WI/D86ePWtmzpxpduzYYTZt2mT69+9vKlasaPbs2XOlLhElGAmfl5k6daqJiooyZcuWNbfffrs5evSou7vkEgsWLDCS8m0rV67Md2xhUzt7470q6J5IMgcPHrQf89Zbb5kGDRqYwMBAU7VqVTN06NCLfoHy9oTmwutLSEgwzZs3N8HBwSY4ONg0btzYvPLKK0V+f2k3c+ZMU61aNRMcHGzatm1rtm/fbm/btWuXadOmjQkICDDVq1c38+bNy/Ped955x9SoUcP4+fmZGjVqmMmTJ9v/UOHJ3nvvPVO2bFnzxx9/5Nlf1M9SYe/3BocPHza9e/c2ERERply5cqZ58+YmISHB3n6pn0W9e/c2VatWNf7+/qZq1aqmd+/eHvPl9lK/gy71eTfm0r+DDhw4YDp16mQCAwNNjRo1zMSJE0vEv4nzLnWPatSoka+tTZs29vffddddJioqyv4zYfDgwSYtLS3POTzpHl3seidMmFBgW9++ffPEuNjPA6vVam655RYTHBxsgoKCTMeOHc3WrVuvzMWhxLMYUwpmsAAAAACAUohpfQAAAADAS5HwAQAAAICXIuEDAAAAAC9FwgcAAAAAXoqEDwAAAAC8FAkfAAAAAHgpEj4AAAAA8FIkfAAAAADgpUj4AMAD9OvXT3369HF3N5xqwYIFqlatmnx8fPTWW2+5uzsexWazqVGjRlq7dq0kadWqVbJYLMrJyXHaOb7//ns1aNBANpvNaTEBACUPCR8AuFDbtm1lsVhksVgUGBio2rVrq1+/ftq6dWue42bNmqVXXnnlkvFycnJksVi0atUqF/XYOc6ePathw4Zp9OjR+vXXX9WrVy93d6lQ0dHRTktIixpryZIlqlixom655RannLcgN910kypVqqRFixa57BwAAM9HwgcALvbII4/o6NGj2r17t958801ZrVbdcMMN+vTTT+3HVKxYURUrVnRjL53ryJEjOnPmjLp27arKlSsrMDAw3zHZ2dlu6JlrFPda5syZo/vvv99FvfmfPn366NVXX3X5eQAAnouEDwBcrFy5coqKilL16tXVtm1bLVy4UPfff78efPBBWa1WSfkf6Zw5c6Zq1aqlgIAARUdH66mnnpIk1alTR5LUrl07WSwW9evXT5L05ptvqkmTJipXrpxq1KihJ554Is/jgefjjx8/XpUqVVKVKlX04osv5unn/v37dccdd6hChQqqWLGiOnTooD/++EOSlJubqyeeeELR0dEqX7682rZtq23bthV4vatWrVKtWrUkSTExMbJYLEpJSVG/fv107733asyYMQoLC1PPnj0lSV988YUaNmyogIAA1alTR++88449VkpKiiwWi5YuXarmzZsrMDBQHTp0UEZGhj744APVrl1bV111lR599FEZYwr93+C9997T1VdfrbJlyyoqKkqDBw+WdK4C++uvv6p///6yWCxq27atJGn58uVq2bKlypcvrypVqmjYsGE6ffq0Pd5TTz2lm2++WTNmzFDVqlXVvHnzQmP9XWpqqtauXasuXboU2t/U1FQ1aNBAQ4YMkTFGb731lqKjo/X++++rVq1aCg4O1sMPP2z/3yU0NFTR0dF6991388Tp2rWrkpKS9OuvvxZ6LgCAdyPhAwA3ePjhh/Xrr79q06ZN+do2bNigCRMm6LXXXtPevXu1ZMkSe6L3ww8/SJI++ugjHT16VLNmzZJ0bkzY888/rx9//FGvvfaa5s2bp9dffz1P3OXLl8tqteqHH37QU089pVGjRtmTtuzsbHXs2FE2m00rV65UcnKyevToodzcXEnSxIkT9fnnn+u9997T5s2bddNNN+nWW2/VyZMn8/X/xhtv1Lp16yRJ69ev19GjR1WtWjVJ0rJly5SVlaXvv/9eL774olJSUhQfH6/4+Hht27ZNjzzyiAYMGKDvv/8+T8xJkybphRde0Lp16/Tzzz/rzjvv1Lvvvqtly5bp3Xff1Zw5c/TZZ58VeK+PHj2q/v37a+LEidq9e7c+++wzNWvWTJK0dOlSVa5cWTNnztTRo0e1dOlSSdKZM2c0btw4bd26Ve+//75WrlypiRMn5om7ZcsWJScn6+uvv9aSJUsKjfV3SUlJioiIUNWqVQtsP3z4sNq0aaO4uDi99tprslgskqSMjAwtWrRIn376qRYvXqzXX39dt912m2w2m9atW6cHH3xQgwYNUnp6uj1W1apVFRUVpaSkpALPBQAoBQwAwGXatGljxo0bl2//mTNnjCTz/vvvG2OM6du3r7n33nuNMcZ8+OGHpl69esZqteZ7n9VqNZLMypUrL3readOmmXbt2tlf9+3b11x77bV5jqlXr555+eWXjTHGzJ8/34SHh5vTp0/ni5WVlWUCAwPN9u3b8+yvW7eu+c9//lPg+ffu3WskmYMHD+bpQ0xMjMnNzbXvGz16tLnhhhvyvLdXr16mZ8+exhhjDh48aCSZxYsX57k2i8VifvvtN/u+Tp06mZEjRxbYl40bN5oKFSqYP//8s8D2qlWrmgULFhTYdt57771natWqZX89YcIEExwcnC9mUWK9+OKLpnHjxnn2rVy50kgye/fuNbVq1TL//ve/87QvWLDAWCwWk5qaat/XqVMnc91119lf5+TkmHLlypnly5fneW/Tpk3Nc889d9E+AQC8FxU+AHAD89fjh+erNxfq0KGDLBaLateuraFDhyohIeGijytK56pGHTt2VNWqVRUcHKynnnpKhw4dynNMgwYN8ryOiopSWlqaJOnHH39UbGysgoKC8sXev3+/srKy1LJlSwUHB9u3/fv368CBA8W67saNG8vH53+/enbv3q2WLVvmOaZVq1bavXt3nn0NGza0/3dkZKTCw8MVERGRZ9+Fla2/n7NRo0aKiYlRv379tGTJEp09e/ai/dy5c6e6d++u6tWrq3z58urfv3+++1m3bl0FBwdf/IILcObMGQUEBBTY1rZtW91+++169tln87WFh4crMjLS/joyMlLXXXed/bWvr69CQ0Pz3YfAwEBlZWUVu58AAO9AwgcAbrBr1y5JUs2aNfO1VaxYUdu2bdOrr74qf39/DRgwQHfccUehsf7880917dpVtWrV0kcffaRNmzZpzJgx9vGB5/n5+eV5bbFY7FP2XyyhPHXqlKRzY/O2bNli33bv3q2HHnqoSNd73t8Tykslsudd2HeLxXLRa/m7MmXKaNWqVVq8eLEiIyP12GOP6cYbb7xo0tetWzdZLBYtXLhQGzdu1EsvvZRvyYSCkuOiCA0N1fHjxwts69y5sz7//HMdPXo0X1tB11yU+/D7778rLCzMob4CAEq+Mu7uAACURi+//LKqVaum66+/vsB2f39/denSRV26dFGfPn3UokULpaWlKTw8XD4+PvaxddK5Ktnx48f1zDPPKCQkRJLyVaMupWHDhlq4cKEyMzPzJTLXXHON/P39dfToUTVv3rx4F3oJV199tRITE/PsW7duna6++mqnnsfX11ft2rVTu3btNGrUKEVGRmrLli2KjY2Vn59fnvt57Ngx7d+/Xx9++KGaNGki6dwyCkXx91gFady4sQ4ePKjs7Ox8lb45c+ZowIABuvXWW7V69WqFhoYW70L/Jjs7WwcOHFDjxo0vKw4AoOSiwgcALnb69Gmlpqbql19+0apVq3Tvvffq3Xff1WuvvaYyZfL/3e2zzz7TK6+8ou3bt+vAgQNavHixwsLCFBoaKovFomrVqikxMVFpaWk6deqUqlevLj8/P82ZM0cHDhzQa6+9pk8++aRYfbznnnsUHBysXr166b///a/27NmjuXPn6tixY6pQoYIeeughPfjgg/roo4908OBBrVu3To8//rh27NhxWffmwQcf1NatW/Xkk09qz549mj17tj788EM98sgjlxX3QsnJyXrmmWe0adMm/fzzz3rnnXcUEBCgGjVqSJJq1KihNWvWKDU1VSdOnNBVV12lq666Sm+88Yb9/s+dO7dI5/p7rIJcf/31Kl++vDZs2JCv7fwi9XXr1lWnTp0KnBSnODZs2KCgoCCnJ+oAgJKDhA8AXGzmzJmqXLmy6tWrpwEDBsjPz08bNmwodFr+kJAQLV68WLfccosaNWqk9evX67PPPpOvr68k6dlnn9XChQtVuXJlPfTQQ4qIiNDrr7+uOXPmqGHDhvr66681ZsyYYvUxICBAX331lWw2m1q3bq0bbrhBS5cutSekzz33nIYNG6b/+7//U/369XXXXXfp0KFDl12BqlGjhj755BN9/PHHatCggWbOnKk333xTN95442XFvVCFChX07bffqmPHjrrmmmv03nvvaenSpfbxcE899ZSSk5NVrVo13XHHHfL19dXChQv19ddf67rrrtPcuXM1adKkIp3r77EK4ufnp3vvvVcffPBBge1lypTR+++/r0qVKqlr167KzMx07MIlffDBB7rnnnvk7+/vcAwAQMlmMUUdQAEAAJwiJSVFLVu21J49e1ShQgWXnOPPP/9UnTp1tG7dOsXExLjkHAAAz0eFDwCAK6xmzZqaMWOGUlJSXHaOn3/+WS+++CLJHgCUclT4AAAAAMBLUeEDAAAAAC9FwgcAAAAAXoqEDwAAAAC8FAkfAAAAAHgpEj4AAAAA8FIkfAAAAADgpUj4AAAAAMBLkfABAAAAgJci4QMAAAAAL/X/WeSPhZJuXPIAAAAASUVORK5CYII=",
+ "text/plain": [
+ "
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/Presentation.qmd b/docs/user-guide/teacher-content/UU-ocean-of-future/Presentation.qmd
new file mode 100644
index 00000000..16777361
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/Presentation.qmd
@@ -0,0 +1,146 @@
+---
+title: "Ocean observations"
+subtitle: "Ocean of the future 13-11-2025" +author: "Emma Daniels & Jamie Atkins
Postdocs @ UU
Virtual Ship Classroom" +format: + revealjs: + slide-number: true + theme: sky + logo: "https://virtualship.readthedocs.io/en/latest/_static/virtual_ship_logo.png" + controls: true + incremental: true +title-slide-attributes: + data-background-image: "https://cloudfront-us-east-2.images.arcpublishing.com/reuters/CQFY2GVMTNJ45KAVHBVEWJYZ44.jpg" + data-background-size: contain + # data-background-iframe: "https://www.youtube.com/embed/qeeipUefe8A?autoplay=1&controls=0&loop=1" + # data-background-video-loop: true + # data-background-video-muted: true +--- + +## + + +## {background-iframe="https://wordwall.net/embed/4f6b5ced54d644c2bab35354305bb0eb?themeId=65&templateId=30&fontStackId=1" background-interactive="true"} + +## Difficulties of Measuring + +- **Vast Scale**: Covers 71% of Earth's surface, average depth 3,688m +- **Real-Time Monitoring**: Difficulty transmitting data from deep or remote locations +- **Extreme conditions**: Pressure, storms and salt require specialized equipment +- **Accessibility**: Remote locations, harsh weather, high operational costs +- **Light Limitation**: Optical methods only work in top ~200m + +## Satellite Observations + + +Note that it's not possible to investigate the interior ocean with satellites + +## Satellite Observations + +::: {.nonincremental} +- **Advantages**: + - Global coverage and accessibility + - Continuous monitoring of large areas + - Cost-effective compared to ship-based surveys + +- **Limitations**: + - Affected by atmospheric conditions + - Cannot penetrate deep into the ocean +::: + +## PACE Satellite (2024) +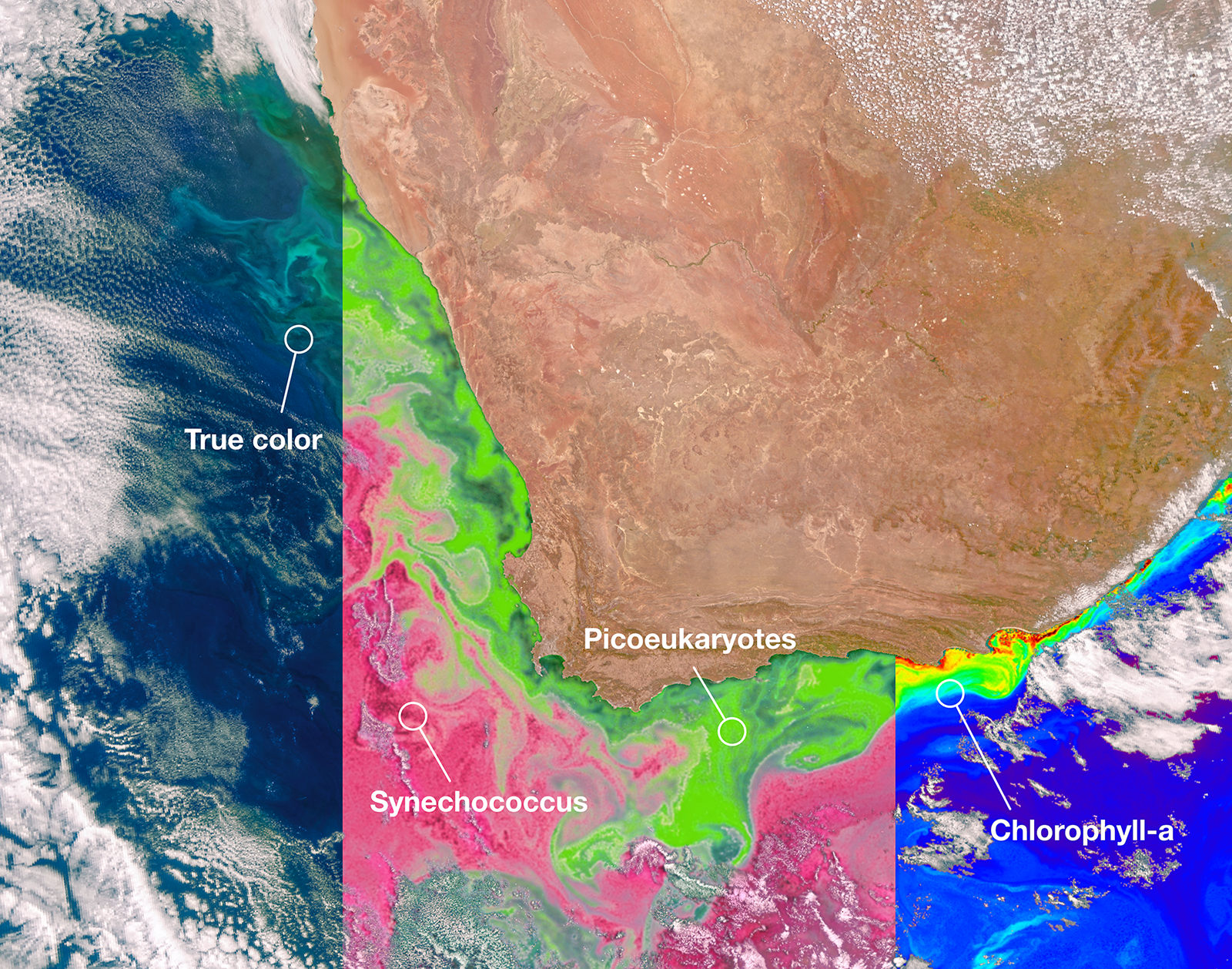 + +## Light Attenuation in the Ocean +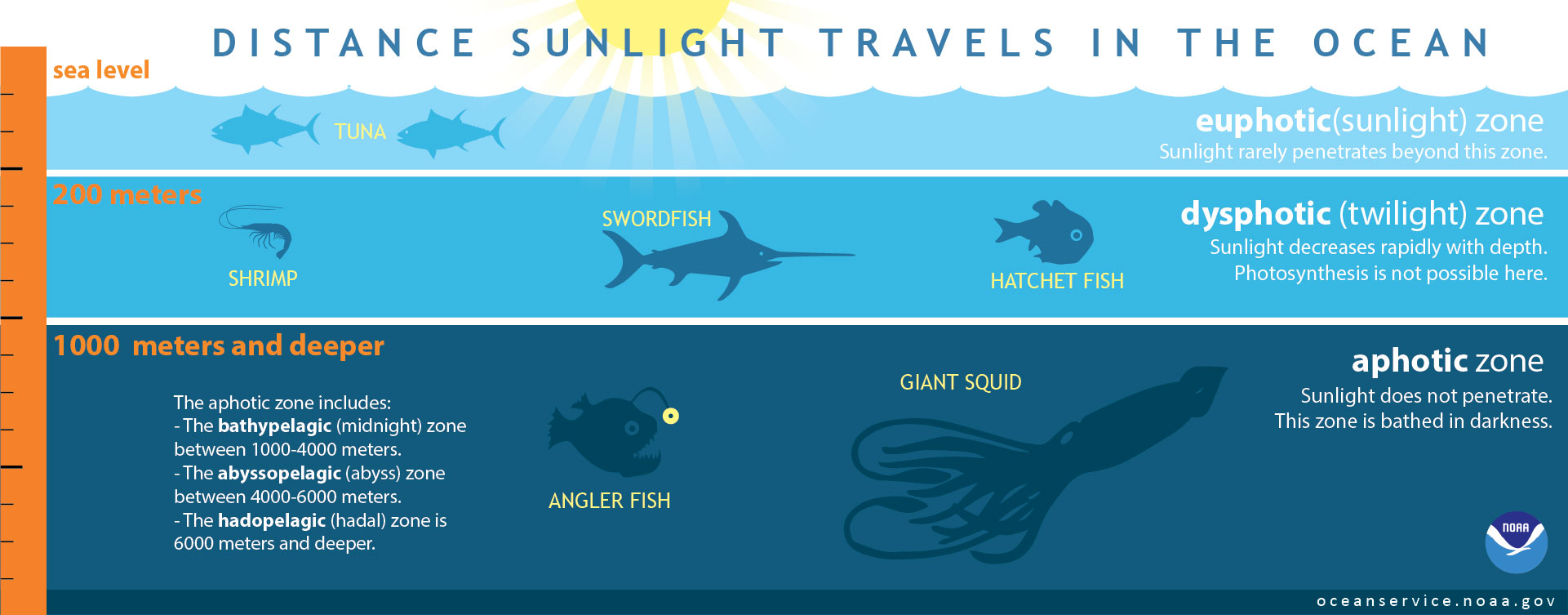 + +## Light Attenuation in the Ocean + +- Remote sensing only observes top few meters of ocean +- Cameras and optical instruments are ineffective at depth +- Alternative methods required: + - Cabled networks and instruments + - Acoustic sensors + - Resurfacing equipment + + + +## Observation Methods + + +## HMS Challenger (1872-1876) + + +## +```{=html} + +``` + +## Ship-Based Measurements + +- **CTD Casts**: Conductivity, Temperature, and Depth profiling and water samples +- **Acoustic Doppler Current Profilers (ADCP)**: Measuring ocean currents +- **Multibeam Echosounders**: High-resolution seafloor mapping +- **Sediment Cores**: Extracting seafloor samples for geological and climate studies +- **Tows**: Biological sampling of plankton and marine organisms + +## + + +## + + +## Benefits of CTD Measurements + +- **High Vertical Resolution**: Continuous profiling from surface to seafloor +- **Targeted Sampling**: Niskin bottles collect water at specific depths of interest +- **Multiple Parameters**: Temperature, salinity, depth, plus additional sensors (O₂, chlorophyll, turbidity) +- **Water Mass Identification**: Characterize ocean layers and circulation patterns +- **Calibration Standard**: Validates satellite and autonomous sensor data + +## +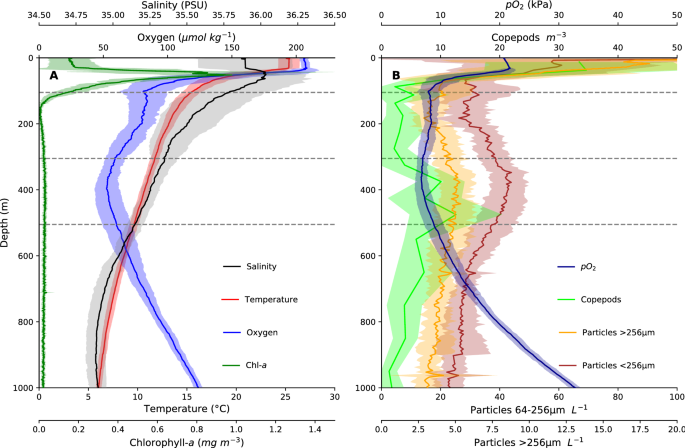 + +## Plankton measurements + +- Water Sampling +- In-Situ Imaging +- Chlorophyll Analysis +- eDNA Filtering +- Tows: nets or CPR + +## + + +## + + +## + + +## + + +## +```{=html} + +``` + +## VirtualShip expedition + +- 9 days of ship time +- Depart and arrive from Texel, Netherlands +- Straight transect starting from the deep shelve +- Inslingeren: (zeem.) opnieuw wennen aan het zeemansleven +- Travel and CTD deployment time +- SURF Research Cloud setup by Jamie +- Instructions in Jupyter Notebook + +## VR and life @ sea + diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial1.ipynb b/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial1.ipynb new file mode 100644 index 00000000..3914f93c --- /dev/null +++ b/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial1.ipynb @@ -0,0 +1,101 @@ +{ + "cells": [ + { + "cell_type": "markdown", + "id": "1536cbb7", + "metadata": {}, + "source": [ + "# Tutorial 1\n", + "\n", + "## VirtualShip exercise\n", + "You can work on the exercise either individually or in pairs. You will work with the same person during the tutorials on November 17th as well. \n", + "You will plan and execute a virtual oceanographic expedition using the VirtualShip software. Follow the steps below to complete the exercise.\n", + "\n", + "### Form and register your duo (preferably with different backgrounds) through Teams\n", + "- Choose a month in the summer half year in which you want to do the expedition (i.e. April - October)\n", + "- Fill in the sheet in the Teams channel and remember your group number (e.g. GROUP1) and the year it is associated with (e.g. 1993)\n", + "\n", + "### Check out your expeditions measurement locations\n", + "https://nioz.marinefacilitiesplanning.com/cruiselocationplanning#\n", + "- Save and upload the cruise track (.xlsx file)\n", + "- Select _Texel - Netherlands_ as the Port of Departure \n", + "- Check out the measurement locations and depths, and travel time between stations\n", + "- Do a (rough) calculation on the time the CTD measurements will take at each station \n", + " - Assume 10 minutes for deployment and retrieval of the CTD at each station\n", + " - Assume 1 m/s for the CTD to go down and up (i.e., for a station at 100 m depth, it will take approximately 200 seconds to go down and up)\n", + "\n", + "### Open a terminal and prepare your expedition in VirtualShip:\n", + "```\n", + "cd data/virtualship_storage/\n", + "virtualship plan GROUP#\n", + "```\n", + "Replace # with your actual group number (e.g., GROUP1)\n", + "\n", + "- Decide on the exact timing of your expedition with in the year and month registered though Teams\n", + "- Remember you have 9 days of ship time available, including travel time from and to Texel, Netherlands\n", + "\n", + "### Follow the instructions in the terminal to set up your expedition\n", + "- From the Schedule Editor select Waypoints & Instrument Selection\n", + "- Click each waypoint to set the time (consider _transit times_ between locations and _time needed for measurements_) \n", + "- Ensure that the CTD and CTD_BGC instruments are selected for use at all waypoints (they should be by default).\n", + "- Save your changes\n", + "\n", + "### Fetch the data needed for your expedition\n", + "```\n", + "virtualship fetch GROUP#\n", + "```\n", + "Replace GROUP# with your actual expedition name (e.g., GROUP1)\n", + "\n", + "- Provide your Copernicus username and password when prompted\n", + "- (Sign up for a Copernicus account if you don't have one yet: https://data.marine.copernicus.eu/register)\n", + "- Practice your patience - a key skill in oceanography! - data download may take some time ;-)\n", + "\n", + "
\n",
+ "In the waiting time you can learn about (life on board) research vessels:\n",
+ "\n",
+ "- https://www.youtube.com/watch?v=hUl0TA-gCK0\n",
+ "\n",
+ "- https://www.youtube.com/watch?v=G82kIgc1imk\n",
+ "\n",
+ "- https://schmidtocean.org/cruise-log-post/four-unexpected-things-i-learned-while-working-on-a-research-vessel/\n",
+ "\n",
+ "Or browse through some blogs from many different cruises, e.g. https://www.nioz.nl/en/blog/topic/1027\n",
+ "
\n",
+ "\n",
+ "\n",
+ "### Start your expedition\n",
+ "`virtualship run GROUP#`\n",
+ "\n",
+ "### Verify your expedition was successful by checking the output directory for data files\n",
+ "- Navigate to the results directory:\n",
+ "`cd GROUP#/results`\n",
+ "- List the files in the results directory:\n",
+ "`ls`\n",
+ "\n",
+ "- Hand in the filepath to your results via Brightspace before Wednesday 19-11-2025 13:00h:\n",
+ "`pwd`"
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.9.6"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial2.ipynb b/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial2.ipynb
new file mode 100644
index 00000000..c73d111f
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/Tutorial2.ipynb
@@ -0,0 +1,55 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "c6b4d243",
+ "metadata": {},
+ "source": [
+ "## Open the notebook CTD transects and run all cells to generate some plots\n",
+ "- modify your data directory\n",
+ "data_dir\n",
+ "\n",
+ "## Plot CTD transects for different variables\n",
+ "Remember your MFP cruise plan and the bathymetry of the North West Shelf TODO: (links)\n",
+ "- Explain why the data is available at different depths throughout the expedition \n",
+ "- Are there temperatures that stand out? For example, temperatures below 0 degrees?\n",
+ "\n",
+ "Play around setting ax.set_ylim() to zoom in on certain depth ranges\n",
+ "- Describe the evolution of salinity along the transect\n",
+ "- Explain why salinity changes close to land\n",
+ "\n",
+ "Oxygen:\n",
+ "- Describe the evolution of oxygen along the transect\n",
+ "- Can you identify any oxygen minimum zones? Hypoxia?\n",
+ "\n",
+ "Nitrate: \n",
+ "Is there nearest to land there are very high nitrate values\n",
+ "- Explain why this is the case\n",
+ "\n",
+ "pH\n",
+ "- Hypothesize why pH is lower in some spots\n",
+ "- Is there an obvious correlation with temperature or other variables?\n",
+ "\n",
+ "Phytoplankton (Chlorophyll)\n",
+ "- Where are the highest concentrations of chlorophyll?\n",
+ "- Is there an obvious correlation with temperature or other variables?\n",
+ "\n",
+ "## Discussion points\n",
+ "- Which variables are most affected by proximity to land?\n",
+ "- How do you expect these variables to change with climate change?\n",
+ "\n",
+ "## Reflection\n",
+ "- Are there locations where you would have liked to take more measurements? \n",
+ "- Why? and how would you modify the cruise plan?\n",
+ "\n"
+ ]
+ }
+ ],
+ "metadata": {
+ "language_info": {
+ "name": "python"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/_publish.yml b/docs/user-guide/teacher-content/UU-ocean-of-future/_publish.yml
new file mode 100644
index 00000000..16810521
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/_publish.yml
@@ -0,0 +1,4 @@
+- source: Presentation.qmd
+ quarto-pub:
+ - id: 79cbaa11-d5f3-4317-895e-162a1a0b3a5f
+ url: https://ammedd.quarto.pub/oceans_of_the_future
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/plot_3D.py b/docs/user-guide/teacher-content/UU-ocean-of-future/plot_3D.py
new file mode 100644
index 00000000..31c5c22b
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/plot_3D.py
@@ -0,0 +1,250 @@
+"""N.B. Quick, inflexible (under active development) version whilst experimenting best approaches!""" # noqa: D400
+# TODO: WORK IN PROGRESS
+
+# %%
+import os
+from glob import glob
+
+import cmocean.cm as cmo
+import matplotlib as mpl
+import numpy as np
+import plotly.graph_objects as go
+import xarray as xr
+
+var = "temperature" # change this to your chosen variable
+
+
+base_dir = os.getcwd()
+filename = "ctd.zarr" if var in ["temperature", "salinity"] else "ctd_bgc.zarr"
+grp_dirs = sorted(glob(os.path.join(base_dir, "GRP????/results/", filename)))
+
+
+VARIABLES = {
+ "temperature": {
+ "cmap": cmo.thermal,
+ "label": "Temperature (°C)",
+ "ds_name": "temperature",
+ },
+ "salinity": {
+ "cmap": cmo.haline,
+ "label": "Salinity (psu)",
+ "ds_name": "salinity",
+ },
+ "oxygen": {
+ "cmap": cmo.oxy,
+ "label": r"Dissolved oxygen (mmol m$^{-3}$)",
+ "ds_name": "o2",
+ },
+ "nitrate": {
+ "cmap": cmo.matter,
+ "label": r"Nitrate (mmol m$^{-3}$)",
+ "ds_name": "no3",
+ },
+ "phosphate": {
+ "cmap": cmo.matter,
+ "label": r"Phosphate (mmol m$^{-3}$)",
+ "ds_name": "po4",
+ },
+ "ph": {
+ "cmap": cmo.balance,
+ "label": "pH",
+ "ds_name": "ph",
+ },
+ "phytoplankton": {
+ "cmap": cmo.algae,
+ "label": r"Total phytoplankton (mmol m$^{-3}$)",
+ "ds_name": "phyc",
+ },
+ "primary_production": {
+ "cmap": cmo.matter,
+ "label": r"Total primary production of phytoplankton (mg m$^{-3}$ day$^{-1}$)",
+ "ds_name": "nppv",
+ },
+ "chlorophyll": {
+ "cmap": cmo.algae,
+ "label": r"Chlorophyll (mg m$^{-3}$)",
+ "ds_name": "chl",
+ },
+}
+
+
+def haversine(lon1, lat1, lon2, lat2):
+ """Great-circle distance (meters) between two points."""
+ lon1, lat1, lon2, lat2 = map(np.radians, [lon1, lat1, lon2, lat2])
+ dlon, dlat = lon2 - lon1, lat2 - lat1
+ a = np.sin(dlat / 2) ** 2 + np.cos(lat1) * np.cos(lat2) * np.sin(dlon / 2) ** 2
+ c = 2 * np.arctan2(np.sqrt(a), np.sqrt(1 - a))
+ return 6371000 * c
+
+
+def distance_from_start(ds):
+ """Add 'distance' variable: meters from first waypoint."""
+ lon0, lat0 = (
+ ds.isel(trajectory=0)["lon"].values[0],
+ ds.isel(trajectory=0)["lat"].values[0],
+ )
+ d = np.zeros_like(ds["lon"].values, dtype=float)
+ for ob, (lon, lat) in enumerate(zip(ds["lon"], ds["lat"], strict=False)):
+ d[ob] = haversine(lon, lat, lon0, lat0)
+ ds["distance"] = xr.DataArray(
+ d,
+ dims=ds["lon"].dims,
+ attrs={"long_name": "distance from first waypoint", "units": "m"},
+ )
+ return ds
+
+
+def descent_only(ds, variable):
+ """Extract descending CTD data (downcast), pad with NaNs for alignment."""
+ min_z_idx = ds["z"].argmin("obs")
+ da_clean = []
+ for i, traj in enumerate(ds["trajectory"].values):
+ idx = min_z_idx.sel(trajectory=traj).item()
+ descent_vals = ds[variable][
+ i, : idx + 1
+ ] # take values from surface to min_z_idx (inclusive)
+ da_clean.append(descent_vals)
+ max_len = max(len(arr[~np.isnan(arr)]) for arr in da_clean)
+ da_padded = np.full((ds["trajectory"].size, max_len), np.nan)
+ for i, arr in enumerate(da_clean):
+ da_dropna = arr[~np.isnan(arr)]
+ da_padded[i, : len(da_dropna)] = da_dropna
+ return xr.DataArray(
+ da_padded,
+ dims=["trajectory", "obs"],
+ coords={"trajectory": ds["trajectory"], "obs": np.arange(max_len)},
+ )
+
+
+def build_masked_array(data_up, profile_indices, n_profiles):
+ arr = np.full((n_profiles, data_up.shape[1]), np.nan)
+ for i, idx in enumerate(profile_indices):
+ if idx is not None:
+ arr[i, :] = data_up.values[idx, :]
+ return arr
+
+
+def get_profile_indices(distance_1d):
+ """
+ Returns regular distance bins and profile indices for CTD transect plotting.
+
+ Bin size is set to one order of magnitude lower than max distance.
+ """
+ dist_min, dist_max = float(distance_1d.min()), float(distance_1d.max())
+ if dist_max > 1e6:
+ dist_step = 1e5
+ elif dist_max > 1e5:
+ dist_step = 1e4
+ elif dist_max > 1e4:
+ dist_step = 1e3
+ else:
+ dist_step = 1e2 # fallback for very short transects
+
+ distance_regular = np.arange(dist_min, dist_max + dist_step, dist_step)
+ threshold = dist_step / 2
+ profile_indices = [
+ np.argmin(np.abs(distance_1d.values - d))
+ if np.min(np.abs(distance_1d.values - d)) < threshold
+ else None
+ for d in distance_regular
+ ]
+ return profile_indices, distance_regular
+
+
+# %%
+
+# pre processing, concat to 3D array
+expeditions = []
+times = []
+for i, path in enumerate(grp_dirs):
+ ctd_ds = xr.open_dataset(path)
+
+ # add distance from start
+ ctd_distance = distance_from_start(ctd_ds)
+
+ # extract descent-only data
+ if i == 0:
+ z_up = descent_only(ctd_distance, "z")
+ d_up = descent_only(ctd_distance, "distance")
+ var_up = descent_only(ctd_distance, VARIABLES[var]["ds_name"])
+
+ # append
+ expeditions.append(var_up)
+ times.append(ctd_ds["time"][0][0].values)
+
+# concat
+var_concat = xr.concat(expeditions, dim="expedition")
+
+
+# 1d array of depth dimension (from deepest trajectory)
+traj_idx, obs_idx = np.where(z_up == np.nanmin(z_up))
+z1d = z_up.values[traj_idx[0], :]
+
+# distance as 1d array
+distance_1d = d_up.isel(obs=0)
+
+# %%
+
+## plotting
+
+# trim to upper 600m
+var_trim = var_concat.where(z_up >= -600)
+
+# Convert cmo.thermal to Plotly colorscale
+thermal_cmap = cmo.thermal
+thermal_colorscale = [
+ [i / 255, mpl.colors.rgb2hex(thermal_cmap(i / 255))] for i in range(256)
+]
+
+# meshgrid for 3D plotting
+expeditions = var_trim["expedition"].values
+trajectories = distance_1d.values
+depths = z1d
+
+xx, yy, zz = np.meshgrid(expeditions, trajectories, depths, indexing="ij")
+
+# values
+values = var_trim.values # shape: (expedition, trajectory, obs)
+valid_values = values[~np.isnan(values)]
+isomin = np.nanpercentile(valid_values, 2.5)
+isomax = np.nanpercentile(valid_values, 97.5)
+
+fig = go.Figure(
+ data=go.Volume(
+ x=xx.flatten(),
+ y=yy.flatten() / 1000.0, # convert to km
+ z=zz.flatten(),
+ value=np.nan_to_num(values, nan=-9999).flatten(),
+ isomin=isomin,
+ isomax=isomax,
+ opacity=0.3,
+ surface_count=21,
+ # opacityscale=[[2, 0.2], [5, 0.5], [5, 0.5], [8, 1]],
+ # opacityscale="extremes",
+ # colorscale=thermal_colorscale,
+ caps=dict(x_show=False, y_show=False, z_show=False), # Hide caps for clarity
+ )
+)
+
+fig.update_layout(
+ scene=dict(
+ zaxis=dict(title="Depth (m)", range=[-600, 0]),
+ yaxis=dict(
+ title="Distance from start (km)",
+ range=[0, np.nanmax(trajectories) / 1000.0],
+ ),
+ xaxis=dict(
+ title="Year",
+ tickvals=np.array([i for i in range(len(expeditions))])[::-1],
+ ticktext=[
+ str(np.datetime64(times[i], "Y")) for i in range(len(expeditions))
+ ][::-1],
+ ),
+ ),
+ margin=dict(l=0, r=0, b=0, t=40),
+ title="3D Volume Plot of " + VARIABLES[var]["label"],
+)
+
+fig.show()
+
+fig.write_html(f"./sample_3D_{var}.html")
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/plot_slider.py b/docs/user-guide/teacher-content/UU-ocean-of-future/plot_slider.py
new file mode 100644
index 00000000..324c5ba4
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/plot_slider.py
@@ -0,0 +1,279 @@
+"""N.B. Quick (under active development) version whilst experimenting best approaches!""" # noqa: D400
+# TODO: WORK IN PROGRESS
+
+# %%
+import os
+from glob import glob
+
+import cmocean.cm as cmo
+import matplotlib as mpl
+import numpy as np
+import plotly.graph_objs as go
+import xarray as xr
+
+var = "primary_production" # change this to your chosen variable
+
+
+base_dir = os.getcwd()
+filename = "ctd.zarr" if var in ["temperature", "salinity"] else "ctd_bgc.zarr"
+grp_dirs = sorted(glob(os.path.join(base_dir, "GRP????/results/", filename)))
+
+
+VARIABLES = {
+ "temperature": {
+ "cmap": cmo.thermal,
+ "label": "Temperature (°C)",
+ "ds_name": "temperature",
+ },
+ "salinity": {
+ "cmap": cmo.haline,
+ "label": "Salinity (PSU)",
+ "ds_name": "salinity",
+ },
+ "oxygen": {
+ "cmap": cmo.oxy,
+ "label": r"Dissolved oxygen (mmol m-3)",
+ "ds_name": "o2",
+ },
+ "nitrate": {
+ "cmap": cmo.matter,
+ "label": r"Nitrate (mmol m-3)",
+ "ds_name": "no3",
+ },
+ "phosphate": {
+ "cmap": cmo.matter,
+ "label": r"Phosphate (mmol m-3)",
+ "ds_name": "po4",
+ },
+ "ph": {
+ "cmap": cmo.balance,
+ "label": "pH",
+ "ds_name": "ph",
+ },
+ "phytoplankton": {
+ "cmap": cmo.algae,
+ "label": r"Total phytoplankton (mmol m-3)",
+ "ds_name": "phyc",
+ },
+ "primary_production": {
+ "cmap": cmo.matter,
+ "label": "Total primary production of phytoplankton (mg m-3 day-1)",
+ "ds_name": "nppv",
+ },
+ "chlorophyll": {
+ "cmap": cmo.algae,
+ "label": "Chlorophyll (mg m-3)",
+ "ds_name": "chl",
+ },
+}
+
+
+def haversine(lon1, lat1, lon2, lat2):
+ """Great-circle distance (meters) between two points."""
+ lon1, lat1, lon2, lat2 = map(np.radians, [lon1, lat1, lon2, lat2])
+ dlon, dlat = lon2 - lon1, lat2 - lat1
+ a = np.sin(dlat / 2) ** 2 + np.cos(lat1) * np.cos(lat2) * np.sin(dlon / 2) ** 2
+ c = 2 * np.arctan2(np.sqrt(a), np.sqrt(1 - a))
+ return 6371000 * c
+
+
+def distance_from_start(ds):
+ """Add 'distance' variable: meters from first waypoint."""
+ lon0, lat0 = (
+ ds.isel(trajectory=0)["lon"].values[0],
+ ds.isel(trajectory=0)["lat"].values[0],
+ )
+ d = np.zeros_like(ds["lon"].values, dtype=float)
+ for ob, (lon, lat) in enumerate(zip(ds["lon"], ds["lat"], strict=False)):
+ d[ob] = haversine(lon, lat, lon0, lat0)
+ ds["distance"] = xr.DataArray(
+ d,
+ dims=ds["lon"].dims,
+ attrs={"long_name": "distance from first waypoint", "units": "m"},
+ )
+ return ds
+
+
+def descent_only(ds, variable):
+ """Extract descending CTD data (downcast), pad with NaNs for alignment."""
+ min_z_idx = ds["z"].argmin("obs")
+ da_clean = []
+ for i, traj in enumerate(ds["trajectory"].values):
+ idx = min_z_idx.sel(trajectory=traj).item()
+ descent_vals = ds[variable][
+ i, : idx + 1
+ ] # take values from surface to min_z_idx (inclusive)
+ da_clean.append(descent_vals)
+ max_len = max(len(arr[~np.isnan(arr)]) for arr in da_clean)
+ da_padded = np.full((ds["trajectory"].size, max_len), np.nan)
+ for i, arr in enumerate(da_clean):
+ da_dropna = arr[~np.isnan(arr)]
+ da_padded[i, : len(da_dropna)] = da_dropna
+ return xr.DataArray(
+ da_padded,
+ dims=["trajectory", "obs"],
+ coords={"trajectory": ds["trajectory"], "obs": np.arange(max_len)},
+ )
+
+
+def build_masked_array(data_up, profile_indices, n_profiles):
+ arr = np.full((n_profiles, data_up.shape[1]), np.nan)
+ for i, idx in enumerate(profile_indices):
+ if idx is not None:
+ arr[i, :] = data_up.values[idx, :]
+ return arr
+
+
+def get_profile_indices(distance_1d):
+ """
+ Returns regular distance bins and profile indices for CTD transect plotting.
+
+ Bin size is set to one order of magnitude lower than max distance.
+ """
+ dist_min, dist_max = float(distance_1d.min()), float(distance_1d.max())
+ if dist_max > 1e6:
+ dist_step = 1e5
+ elif dist_max > 1e5:
+ dist_step = 1e4
+ elif dist_max > 1e4:
+ dist_step = 1e3
+ else:
+ dist_step = 1e2 # fallback for very short transects
+
+ distance_regular = np.arange(dist_min, dist_max + dist_step, dist_step)
+ threshold = dist_step / 2
+ profile_indices = [
+ np.argmin(np.abs(distance_1d.values - d))
+ if np.min(np.abs(distance_1d.values - d)) < threshold
+ else None
+ for d in distance_regular
+ ]
+ return profile_indices, distance_regular
+
+
+# %%
+
+# pre processing, concat to 3D array
+expeditions = []
+times = []
+for i, path in enumerate(grp_dirs):
+ ctd_ds = xr.open_dataset(path)
+
+ # add distance from start
+ ctd_distance = distance_from_start(ctd_ds)
+
+ # extract descent-only data
+ if i == 0:
+ z_up = descent_only(ctd_distance, "z")
+ d_up = descent_only(ctd_distance, "distance")
+ var_up = descent_only(ctd_distance, VARIABLES[var]["ds_name"])
+
+ # append
+ expeditions.append(var_up)
+ times.append(ctd_ds["time"][0][0].values)
+
+# concat
+var_concat = xr.concat(expeditions, dim="expedition")
+var_concat["expedition"] = times
+
+# 1d array of depth dimension (from deepest trajectory)
+traj_idx, obs_idx = np.where(z_up == np.nanmin(z_up))
+z1d = z_up.values[traj_idx[0], :]
+
+# distance as 1d array
+distance_1d = d_up.isel(obs=0)
+
+# %%
+
+## plotting (interactive with Plotly)
+
+depth_lim = -200 # [m]
+
+# trim to upper 600m
+var_trim = var_concat.where(z_up >= depth_lim)
+
+
+# Prepare colorscale for Plotly from matplotlib colormap
+def mpl_to_plotly(cmap, n=256):
+ return [[i / (n - 1), mpl.colors.rgb2hex(cmap(i / (n - 1)))] for i in range(n)]
+
+
+plotly_cmap = mpl_to_plotly(VARIABLES[var]["cmap"])
+
+# Prepare slider steps
+steps = []
+data = []
+for t in range(var_trim.shape[0]):
+ seabed = xr.where(np.isnan(var_trim[t]), 1, None).T
+
+ # main cross-section
+ trace = go.Heatmap(
+ z=var_trim[t].T,
+ x=distance_1d / 1000.0, # distance in km
+ y=z1d,
+ zmin=np.nanmin(var_trim.values),
+ zmax=np.nanmax(var_trim.values),
+ colorscale=plotly_cmap,
+ colorbar=dict(title=VARIABLES[var]["label"]),
+ showscale=True,
+ visible=(t == 0),
+ customdata=None,
+ hovertemplate="Distance: %{x:.2f} kmDepth: %{z:.1f} m
Value: %{value:.2f}
\n",
+ "This script uses an infinite `while` loop to refresh the plotting indefinitely. To stop the running, the \"Interupt the kernel\" button (square button) should be pressed.\n",
+ "
\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "64a8edb1-46c3-488f-a0dc-d104d47bbaa5",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import time\n",
+ "import os\n",
+ "import sys\n",
+ "import yaml\n",
+ "import numpy as np\n",
+ "from pathlib import Path\n",
+ "from matplotlib import pyplot as plt\n",
+ "from IPython.display import clear_output"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 2,
+ "id": "d93d52a2-b55d-43b6-bc2e-a56aecbe8fa7",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# plot refresh rate\n",
+ "REFRESH = 30 # [seconds]"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 3,
+ "id": "2c61c6b5-e729-46d0-bc9e-2f87a447ee76",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# config\n",
+ "BASE_DIR = Path(\"/home/shared/data/virtualship_storage/\")\n",
+ "SAIL_OUT_TIME = np.timedelta64(3, \"D\") # [days]\n",
+ "CTD_TIME = np.timedelta64(200, \"m\") # [minutes]\n",
+ "SHIP_TIME_THRESHOLD = 9 # [days]\n",
+ "ROTATION_CHANGE = 18 # when to change from 45 to 90 degree rotation in x axis labels"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 4,
+ "id": "4a91fae1-46d1-4e49-855a-b0bec7b127d9",
+ "metadata": {
+ "jupyter": {
+ "source_hidden": true
+ }
+ },
+ "outputs": [],
+ "source": [
+ "def preprocess(\n",
+ " base_dir: Path, sail_out_time: np.timedelta64, ctd_time: np.timedelta64\n",
+ ") -> dict:\n",
+ " \"\"\"\n",
+ " Reads schedule data from YAML files, calculates the total expedition duration\n",
+ " for each group, and returns a dictionary of valid groups and their durations.\n",
+ " \"\"\"\n",
+ " groups = {}\n",
+ "\n",
+ " # group directories 1 to 66 (inclusive)\n",
+ " for i in range(1, 67):\n",
+ " group_name = f\"GROUP{i}\"\n",
+ " group_dir = base_dir / group_name\n",
+ " schedule_file = group_dir / \"schedule.yaml\"\n",
+ "\n",
+ " if not schedule_file.exists():\n",
+ " groups[group_name] = np.nan\n",
+ " continue\n",
+ "\n",
+ " try:\n",
+ " with open(schedule_file, \"r\", encoding=\"utf-8\") as f:\n",
+ " schedule_data = yaml.safe_load(f)\n",
+ "\n",
+ " waypoints = schedule_data.get(\"waypoints\", [])\n",
+ "\n",
+ " if not waypoints:\n",
+ " groups[group_name] = np.nan\n",
+ " continue\n",
+ "\n",
+ " start_time_str = waypoints[0].get(\"time\")\n",
+ " end_time_str = waypoints[-1].get(\"time\")\n",
+ "\n",
+ " if not start_time_str or not end_time_str:\n",
+ " groups[group_name] = np.nan\n",
+ " else:\n",
+ " start_time, end_time = (\n",
+ " np.datetime64(start_time_str),\n",
+ " np.datetime64(end_time_str),\n",
+ " )\n",
+ "\n",
+ " difference = end_time - start_time\n",
+ " difference_days = difference.astype(\"timedelta64[D]\") # [days]\n",
+ "\n",
+ " total_time_timedelta = difference_days + sail_out_time + ctd_time\n",
+ " total_time_days = total_time_timedelta.astype(\n",
+ " \"timedelta64[h]\"\n",
+ " ) / np.timedelta64(24, \"h\")\n",
+ "\n",
+ " groups[group_name] = total_time_days.item()\n",
+ "\n",
+ " except Exception as e:\n",
+ " groups[group_name] = np.nan\n",
+ " print(f\"Error processing {group_name}: {e}\", file=sys.stderr)\n",
+ "\n",
+ " # filter dict to remove groups with NaN\n",
+ " filter_groups = {key: value for key, value in groups.items() if not np.isnan(value)}\n",
+ "\n",
+ " return filter_groups"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "786e2cb8-1dc0-46c0-a454-37ac9e603329",
+ "metadata": {
+ "jupyter": {
+ "source_hidden": true
+ }
+ },
+ "outputs": [],
+ "source": [
+ "def plot(filter_groups: dict, ship_time_threshold: float, rotation_change: int):\n",
+ " \"\"\"\n",
+ " Generates a bar plot showing the expedition duration for each group\n",
+ " relative to the ship time threshold.\n",
+ " \"\"\"\n",
+ " groups_keys = list(filter_groups.keys())\n",
+ " groups_values = list(filter_groups.values())\n",
+ "\n",
+ " # bar colors dependent on whether above or below ship time threshold\n",
+ " bar_colors = [\n",
+ " \"crimson\" if value > ship_time_threshold else \"mediumseagreen\"\n",
+ " for value in groups_values\n",
+ " ]\n",
+ "\n",
+ " # fig\n",
+ " fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(16, 8), dpi=300)\n",
+ "\n",
+ " # bars\n",
+ " ax.bar(\n",
+ " groups_keys,\n",
+ " groups_values,\n",
+ " color=bar_colors,\n",
+ " edgecolor=\"k\",\n",
+ " linewidth=1.5,\n",
+ " zorder=3,\n",
+ " width=0.7,\n",
+ " )\n",
+ "\n",
+ " # labels and title\n",
+ " ax.set_ylabel(\"Days\", fontsize=15)\n",
+ " ax.set_title(\"Expedition Duration\", fontsize=20)\n",
+ "\n",
+ " # customise ticks\n",
+ " ax.set_xticks(ax.get_xticks())\n",
+ " rotation = 45 if len(groups_values) <= rotation_change else 90\n",
+ " ax.set_xticklabels(groups_keys, rotation=rotation, ha=\"center\", fontsize=15)\n",
+ " ax.tick_params(axis=\"y\", labelsize=15)\n",
+ "\n",
+ " # set y-limit based on the maximum valid value\n",
+ " max_duration = np.nanmax(groups_values) if groups_values else 0\n",
+ " ax.set_ylim(0, max_duration + 0.5)\n",
+ "\n",
+ " # grid\n",
+ " ax.set_facecolor(\"gainsboro\")\n",
+ " ax.grid(axis=\"y\", linestyle=\"-\", alpha=1.0, color=\"white\")\n",
+ "\n",
+ " # horizontal line for threshold days\n",
+ " ax.axhline(\n",
+ " y=ship_time_threshold,\n",
+ " color=\"r\",\n",
+ " linestyle=\"--\",\n",
+ " linewidth=2.5,\n",
+ " label=\"Ship-time limit\",\n",
+ " zorder=2,\n",
+ " )\n",
+ "\n",
+ " plt.legend(fontsize=15, loc=\"upper left\")\n",
+ " plt.tight_layout()\n",
+ " plt.show()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 6,
+ "id": "73a78ef2-1b58-48be-802f-0a447c9549a2",
+ "metadata": {
+ "jupyter": {
+ "source_hidden": true
+ }
+ },
+ "outputs": [],
+ "source": [
+ "def periodic_task(interval_seconds: int):\n",
+ " \"\"\"\n",
+ " Loop that runs the preprocessing and plotting functions periodically.\n",
+ " :param interval_seconds: The number of seconds to wait between runs.\n",
+ " \"\"\"\n",
+ " while True:\n",
+ " try:\n",
+ " clear_output(wait=True)\n",
+ " print(f\"--- Running task at {time.ctime()} ---\")\n",
+ "\n",
+ " data = preprocess(BASE_DIR, SAIL_OUT_TIME, CTD_TIME)\n",
+ "\n",
+ " if data:\n",
+ " plot(data, SHIP_TIME_THRESHOLD, ROTATION_CHANGE)\n",
+ " else:\n",
+ " print(\"No valid data found to plot.\")\n",
+ "\n",
+ " except KeyboardInterrupt:\n",
+ " print(\"\\nPeriodic task interrupted by user.\")\n",
+ " break\n",
+ " except Exception as e:\n",
+ " print(f\"\\nAn error occurred during the periodic run: {e}\")\n",
+ "\n",
+ " time.sleep(interval_seconds)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 7,
+ "id": "939a0a6b-9261-4e34-80f0-168e9bb133dd",
+ "metadata": {
+ "collapsed": true,
+ "jupyter": {
+ "outputs_hidden": true
+ },
+ "scrolled": true
+ },
+ "outputs": [
+ {
+ "name": "stdout",
+ "output_type": "stream",
+ "text": [
+ "--- Running task at Wed Nov 12 21:45:02 2025 ---\n",
+ "No valid data found to plot.\n"
+ ]
+ },
+ {
+ "ename": "KeyboardInterrupt",
+ "evalue": "",
+ "output_type": "error",
+ "traceback": [
+ "\u001b[31m---------------------------------------------------------------------------\u001b[39m",
+ "\u001b[31mKeyboardInterrupt\u001b[39m Traceback (most recent call last)",
+ "\u001b[36mCell\u001b[39m\u001b[36m \u001b[39m\u001b[32mIn[7]\u001b[39m\u001b[32m, line 1\u001b[39m\n\u001b[32m----> \u001b[39m\u001b[32m1\u001b[39m \u001b[43mperiodic_task\u001b[49m\u001b[43m(\u001b[49m\u001b[43minterval_seconds\u001b[49m\u001b[43m \u001b[49m\u001b[43m=\u001b[49m\u001b[43m \u001b[49m\u001b[43mREFRESH\u001b[49m\u001b[43m)\u001b[49m\n",
+ "\u001b[36mCell\u001b[39m\u001b[36m \u001b[39m\u001b[32mIn[6]\u001b[39m\u001b[32m, line 24\u001b[39m, in \u001b[36mperiodic_task\u001b[39m\u001b[34m(interval_seconds)\u001b[39m\n\u001b[32m 21\u001b[39m \u001b[38;5;28;01mexcept\u001b[39;00m \u001b[38;5;167;01mException\u001b[39;00m \u001b[38;5;28;01mas\u001b[39;00m e:\n\u001b[32m 22\u001b[39m \u001b[38;5;28mprint\u001b[39m(\u001b[33mf\u001b[39m\u001b[33m\"\u001b[39m\u001b[38;5;130;01m\\n\u001b[39;00m\u001b[33mAn error occurred during the periodic run: \u001b[39m\u001b[38;5;132;01m{\u001b[39;00me\u001b[38;5;132;01m}\u001b[39;00m\u001b[33m\"\u001b[39m)\n\u001b[32m---> \u001b[39m\u001b[32m24\u001b[39m \u001b[43mtime\u001b[49m\u001b[43m.\u001b[49m\u001b[43msleep\u001b[49m\u001b[43m(\u001b[49m\u001b[43minterval_seconds\u001b[49m\u001b[43m)\u001b[49m\n",
+ "\u001b[31mKeyboardInterrupt\u001b[39m: "
+ ]
+ }
+ ],
+ "source": [
+ "periodic_task(interval_seconds=REFRESH)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "be0a7bfd-dc2a-437a-8fa6-ffb78cdd2687",
+ "metadata": {},
+ "outputs": [],
+ "source": []
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "virtualship",
+ "language": "python",
+ "name": "virtualship"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.12.12"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/docs/user-guide/teacher-content/UU-ocean-of-future/timeseries.py b/docs/user-guide/teacher-content/UU-ocean-of-future/timeseries.py
new file mode 100644
index 00000000..24d97547
--- /dev/null
+++ b/docs/user-guide/teacher-content/UU-ocean-of-future/timeseries.py
@@ -0,0 +1,177 @@
+# %%
+
+"""N.B. Quick, inflexible (under active development) version whilst experimenting best approaches!""" # noqa: D400
+# TODO: WORK IN PROGRESS!
+
+import glob
+import os
+
+import matplotlib.pyplot as plt
+import numpy as np
+import xarray as xr
+
+# TODO: incorporate uncertainty estimates in the plots, box plots per year/expedition
+
+# TODO: build conmplexity of plots, single points -> lines -> uncertainty/boxplots -> colour boxplots by month of the (half) year
+
+# TODO: timeseries - can just do it for surface...
+
+# TODO: 3D plots of CTD Transects!
+
+variables = ["phyc", "temperature", "salinity", "o2", "no3", "po4"]
+
+base_dir = os.getcwd()
+
+dict_vars = {}
+for var in variables:
+ print(f"Processing variable: {var}")
+ filename = "ctd.zarr" if var in ["temperature", "salinity"] else "ctd_bgc.zarr"
+ grp_dirs = sorted(glob.glob(os.path.join(base_dir, "GRP????/results/", filename)))
+
+ var_values = []
+ times = []
+
+ tmp = {}
+ for zarr_path in grp_dirs:
+ ds = xr.open_zarr(zarr_path)
+
+ # extract variable values and time
+ var_values.append(ds[var].values.flatten())
+ times.append(ds["time"].values[0][0])
+
+ # organise to dict
+ tmp["values"], tmp["time"] = var_values, times
+
+ # master dict
+ dict_vars[var] = tmp
+
+# %%
+
+plot_dict = {
+ "phyc": {
+ "label": "Phytoplankton",
+ "units": "mmol m$^{-3}$",
+ "color": "forestgreen",
+ },
+ "temperature": {
+ "label": "Temperature",
+ "units": "°C",
+ "color": "crimson",
+ },
+ "salinity": {
+ "label": "Salinity",
+ "units": "PSU",
+ "color": "lightseagreen",
+ },
+ "o2": {
+ "label": "Oxygen",
+ "units": "mmol m$^{-3}$",
+ "color": "dodgerblue",
+ },
+ "no3": {
+ "label": "Nitrate",
+ "units": "mmol m$^{-3}$",
+ "color": "darkorchid",
+ },
+ "po4": {
+ "label": "Phosphate",
+ "units": "mmol m$^{-3}$",
+ "color": "coral",
+ },
+}
+
+# %%
+
+combined_vars = [v for v in variables if v not in ["no3", "po4"]] + ["no3_po4"]
+fig, axs = plt.subplots(
+ len(combined_vars), 1, figsize=(10, 10), dpi=96, sharex=True, sharey=False
+)
+
+for i, ax in enumerate(axs):
+ if i < len(combined_vars) - 1:
+ var = combined_vars[i]
+ color = plot_dict[var]["color"]
+
+ ax.scatter(
+ dict_vars[var]["time"],
+ [np.nanmean(values) for values in dict_vars[var]["values"]],
+ color=color,
+ zorder=3,
+ s=50,
+ )
+
+ ax.plot(
+ dict_vars[var]["time"],
+ [np.nanmean(values) for values in dict_vars[var]["values"]],
+ linestyle="dotted",
+ alpha=1.0,
+ color=color,
+ lw=2.25,
+ )
+
+ ax.set_title(f"{plot_dict[var]['label']}", fontsize=12)
+ ax.set_ylabel(plot_dict[var]["units"], fontsize=11)
+ else:
+ color_no3 = plot_dict["no3"]["color"]
+ color_po4 = plot_dict["po4"]["color"]
+
+ ax.scatter(
+ dict_vars["no3"]["time"],
+ [np.nanmean(values) for values in dict_vars["no3"]["values"]],
+ color=color_no3,
+ zorder=3,
+ s=50,
+ label=plot_dict["no3"]["label"],
+ )
+ ax.plot(
+ dict_vars["no3"]["time"],
+ [np.nanmean(values) for values in dict_vars["no3"]["values"]],
+ linestyle="dotted",
+ alpha=1.0,
+ color=color_no3,
+ lw=2.25,
+ )
+ ax.set_ylabel(plot_dict["no3"]["units"], fontsize=11, color=color_no3)
+ ax.tick_params(axis="y", labelcolor=color_no3)
+
+ ax2 = ax.twinx()
+ ax2.scatter(
+ dict_vars["po4"]["time"],
+ [np.nanmean(values) for values in dict_vars["po4"]["values"]],
+ color=color_po4,
+ zorder=3,
+ s=50,
+ label=plot_dict["po4"]["label"],
+ )
+ ax2.plot(
+ dict_vars["po4"]["time"],
+ [np.nanmean(values) for values in dict_vars["po4"]["values"]],
+ linestyle="dotted",
+ alpha=1.0,
+ color=color_po4,
+ lw=2.25,
+ )
+ ax2.set_ylabel(plot_dict["po4"]["units"], fontsize=11, color=color_po4)
+ ax2.tick_params(axis="y", labelcolor=color_po4)
+
+ ax.set_title("Nutrients", fontsize=12)
+
+ handles, labels = [], []
+ for a in [ax, ax2]:
+ h, label = a.get_legend_handles_labels()
+ handles += h
+ labels += label
+ ax.legend(handles, labels, loc="upper right")
+
+ ax.set_xlim(np.datetime64("1993-01-01"), np.datetime64("2025-12-31"))
+
+ if i == len(axs) - 1: # bottom panel only for single column of subplots
+ ax.set_xlabel("Time")
+
+ ax.set_facecolor("gainsboro")
+ ax.grid(color="white", linewidth=1.0)
+
+
+plt.tight_layout()
+plt.show()
+# %%
diff --git a/docs/user-guide/teacher-content/index.md b/docs/user-guide/teacher-content/index.md
index d3b6f5c8..864392a5 100644
--- a/docs/user-guide/teacher-content/index.md
+++ b/docs/user-guide/teacher-content/index.md
@@ -12,7 +12,7 @@ The 360 videos are available on our YouTube channel [**_@VirtualShip Classroom_*
- Smartphones/Tablets – Move your device or swipe the screen to explore.
- PC/Mac Browsers – Click and drag with your mouse to look around.
-The VSC design focuses on creating didactically sound, authentic learning experiences grounded in established learning theories in science education, such as constructivism [(Piaget 1954)](https://doi.org/10.4324/9781315009650) and constructionism [(Papert 1980)](https://worrydream.com/refs/Papert_1980_-_Mindstorms,_1st_ed.pdf). By integrating realistic tasks and a gamified narrative approach within Jupyter notebooks, students learn within a digital replica of the real world, constructing knowledge through ‘learning by doing’ and ‘trial and error’ as they explore oceanography concepts, research methods, and analysis tools.
+The VSC design focuses on creating didactically sound, authentic learning experiences grounded in established learning theories in science education, such as constructivism [(Piaget 1954)](https://www.taylorfrancis.com/books/mono/10.4324/9781315009650/construction-reality-child-jean-piaget) and constructionism [(Papert 1980)](https://worrydream.com/refs/Papert_1980_-_Mindstorms,_1st_ed.pdf). By integrating realistic tasks and a gamified narrative approach within Jupyter notebooks, students learn within a digital replica of the real world, constructing knowledge through ‘learning by doing’ and ‘trial and error’ as they explore oceanography concepts, research methods, and analysis tools.
We evaluated in several (under)graduate courses and find that the VirtualShip Classroom is highly engaging, and students report on enhanced confidence and knowledge [(Daniels et al. 2025)](https://current-journal.com/articles/10.5334/cjme.121).
@@ -24,3 +24,12 @@ caption: Teaching material
ILOs.ipynb
```
+
+## UU Ocean of the Future
+
+```{nbgallery}
+---
+maxdepth: 1
+---
+UU-ocean-of-future/Tutorial1.ipynb
+```
diff --git a/docs/user-guide/tutorials/index.md b/docs/user-guide/tutorials/index.md
index 9b181aff..592aba69 100644
--- a/docs/user-guide/tutorials/index.md
+++ b/docs/user-guide/tutorials/index.md
@@ -6,6 +6,7 @@ maxdepth: 1
caption: Tutorials
---
surf_research_cloud_setup.ipynb
+surf_collaborative_setup.ipynb
ADCP_data_tutorial.ipynb
CTD_data_tutorial.ipynb
Drifter_data_tutorial.ipynb
diff --git a/docs/user-guide/tutorials/surf_collaborative_setup.ipynb b/docs/user-guide/tutorials/surf_collaborative_setup.ipynb
new file mode 100644
index 00000000..94bb1e43
--- /dev/null
+++ b/docs/user-guide/tutorials/surf_collaborative_setup.ipynb
@@ -0,0 +1,120 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "98770716",
+ "metadata": {},
+ "source": [
+ "# SURF Resarch Cloud: Collaborative Workspace Setup Guide\n",
+ "\n",
+ "```\n",
+ "Note: This guide is specific to students who are enrolled at Utrecht University.\n",
+ "```\n",
+ "\n",
+ "In the class, we will use VirtualShip in the cloud (in this case, SURF Research Cloud - called SURF RC from here-on). This has several advantages:\n",
+ "\n",
+ "- You aren't limited to the power of your laptop.\n",
+ "- Everyone can work in the same collaborative environment.\n",
+ "- The environment is pre-configured with all necessary software and dependencies.\n",
+ "\n",
+ "\n",
+ "## 1. Accepting SURF RC invite\n",
+ "\n",
+ "In your student email you'll have an invite from SURF Research Access Management (SRAM) to join a project on SURF RC. Accept this invite.\n",
+ "\n",
+ "## 2. Open the workspace\n",
+ "\n",
+ "Navigate to the [SURF Research Cloud Dashboard](https://portal.live.surfresearchcloud.nl/), and click \"access\" on the shared workspace.\n",
+ "\n",
+ "\n",
+ "## 3. Open Terminal session\n",
+ "\n",
+ "In the Jupyter launcher, you should see an option to open Terminal session. Click this to open Terminal.\n",
+ "\n",
+ "\n",
+ "## 4. Launch VirtualShip environment\n",
+ "\n",
+ "❗️ Before proceeding any further, you should type the following command in the Terminal and then hit Enter: `bash` ❗️\n",
+ "\n",
+ "\n",
+ "This will ensure that you are in a bash shell for the rest of the setup.\n",
+ "\n",
+ "You will see that the Terminal prompt has changed to something like:\n",
+ "\n",
+ "```bash\n",
+ "(base) metheuser@mywsp:\n",
+ "```\n",
+ "\n",
+ "This is conda telling you that you are currently in the \"base\" environment.\n",
+ "\n",
+ "From here, you already have another environment set up for you. Running `conda env list` in the Terminal, you should see:\n",
+ "\n",
+ "```bash\n",
+ "conda env list\n",
+ "\n",
+ "# conda environments:\n",
+ "#\n",
+ "base * /etc/miniconda3\n",
+ "virtualship /etc/miniconda3/envs/virtualship\n",
+ "```\n",
+ "\n",
+ "Next, to launch the VirtualShip environment, type the following command in the Terminal and hit Enter: `conda activate virtualship`\n",
+ "\n",
+ "This will activate the VirtualShip conda environment, which has all the necessary dependencies installed. You can confirm that you are in the correct environment by checking that your Terminal prompt now looks something like:\n",
+ "\n",
+ "```bash\n",
+ "(virtualship) metheuser@mywsp:\n",
+ "```\n",
+ "\n",
+ "With the `virtualship` environment, you now have access to the `virtualship` command in your Terminal.\n",
+ "\n",
+ "This can be confirmed by typing the following command in the Terminal and hitting Enter: `virtualship --help`.\n",
+ "\n",
+ "You should see something like:\n",
+ "\n",
+ "```\n",
+ "virtualship --help\n",
+ "\n",
+ "Usage: virtualship [OPTIONS] COMMAND [ARGS]...\n",
+ "\n",
+ "Options:\n",
+ " --version Show the version and exit.\n",
+ " --help Show this message and exit.\n",
+ "\n",
+ "Commands:\n",
+ " fetch Download input data for an expedition.\n",
+ " init Initialize a directory for a new expedition, with an example...\n",
+ " plan Launch UI to help build schedule and ship config files.\n",
+ " run Run the expedition.\n",
+ " ```\n",
+ "\n",
+ "\n",
+ "If you close the Terminal window at any point, you may need to re-open it and re-run the `bash` and `conda activate virtualship` commands to ensure you are in a bash shell and the correct conda environment. \n",
+ "
\n",
+ "\n",
+ "## 5. Navigate to the shared storage folder\n",
+ "\n",
+ "We will be working from a shared storage folder in the workspace. To navigate to this folder, type the following command in the Terminal and hit Enter: `cd data/virtualship_storage/`\n",
+ "\n",
+ "Depending on the specific set-up for the course you are taking, if you now enter `ls` to list the contents of the directory, you may see various group folders, for example: `GROUP1`, `GROUP2`, ... and so on.\n",
+ "\n",
+ "If you have been pre-assigned into groups, this is where will be working from for the rest of the course, with the group name for each folder corresponding to your assigned group.\n",
+ "\n",
+ "You are ready to continue with the VirtualShip analysis workflow as described in the course materials and/or VirtualShip [quickstart guide](https://virtualship.readthedocs.io/en/latest/user-guide/quickstart.html)."
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "3926833e",
+ "metadata": {},
+ "source": []
+ }
+ ],
+ "metadata": {
+ "language_info": {
+ "name": "python"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
 +
+  + Note that it’s not possible to investigate the interior ocean with
+ satellites
+
+ Note that it’s not possible to investigate the interior ocean with
+ satellites
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+  +
+