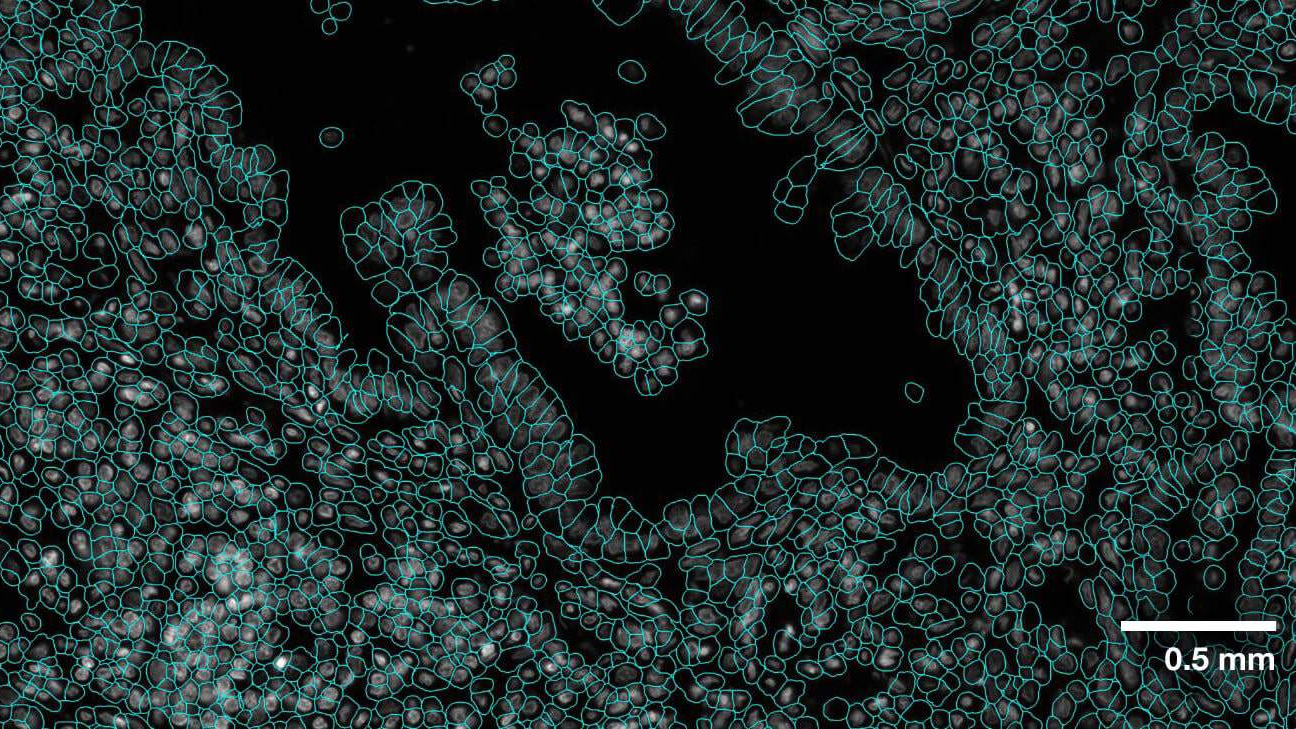
The VISTA2D is a cell segmentation training and inference pipeline for cell imaging [Blog].
A pretrained model was trained on collection of 15K public microscopy images. The data collection and training can be reproduced following the tutorial. Alternatively, the model can be retrained on your own dataset. The pretrained vista2d model achieves good performance on diverse set of cell types, microscopy image modalities, and can be further finetuned if necessary. The codebase utilizes several components from other great works including SegmentAnything and Cellpose, which must be pip installed as dependencies. Vista2D codebase follows MONAI bundle format and its specifications.
- Robust deep learning algorithm based on transformers
- Generalist model as compared to specialist models
- Multiple dataset sources and file formats supported
- Multiple modalities of imaging data collectively supported
- Multi-GPU and multinode training support
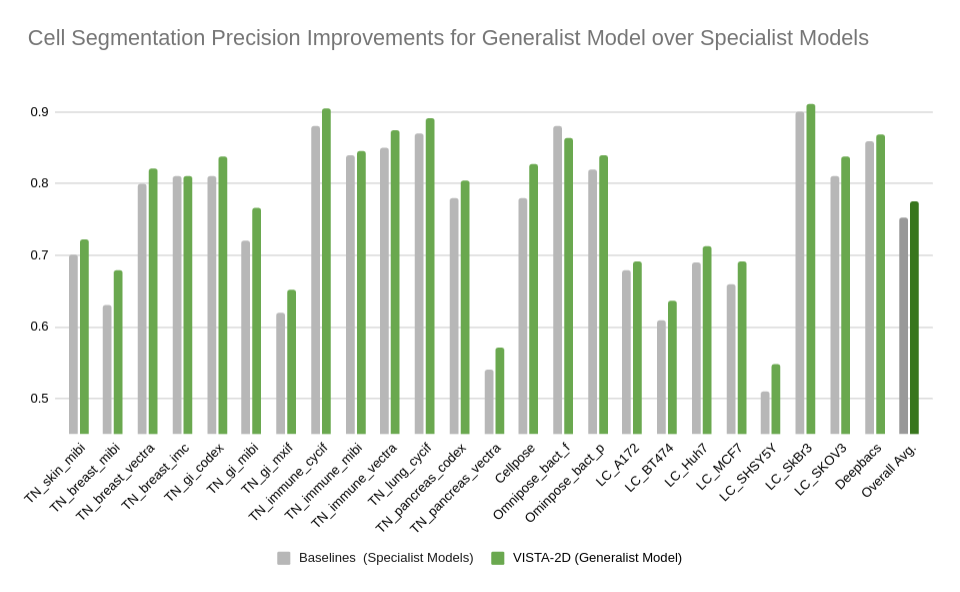
Evaluation was performed for the VISTA2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and more. A total of ~15K annotated cell images were collected to train the generalist VISTA2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types. The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. Average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA2D model trained only on a particular dataset or a subset of data.
pip install monai fire tifffile imagecodecs pillow fastremap
pip install --no-deps cellpose natsort roifile
pip install git+https://github.com/facebookresearch/segment-anything.git
pip install mlflow psutil pynvml #optional for MLFlow support
python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yamlpython -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --quick True --train#trainer#max_epochs 3torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yamlpython -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --pretrained_ckpt_name model.pt --mode eval(can append --quick True for quick demoing)
torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode evalpython -m monai.bundle run --config_file configs/inference.jsontorchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode infer --pretrained_ckpt_name model.pt(can append --quick True for quick demoing)
(we use a smaller learning rate, small number of epochs, and initialize from a checkpoint)
python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --learning_rate=0.001 --train#trainer#max_epochs 20 --pretrained_ckpt_path /path/to/saved/model.ptTo disable the segmentation writing:
--postprocessing []
Load a checkpoint for validation or inference (relative path within results directory):
--pretrained_ckpt_name "model.pt"
Load a checkpoint for validation or inference (absolute path):
--pretrained_ckpt_path "/path/to/another/location/model.pt"
--mode eval or --mode inferwill use the corresponding configurations from the validate or infer
of the configs/hyper_parameters.yaml.
By default the generated model.pt corresponds to the checkpoint at the best validation score,
model_final.pt is the checkpoint after the latest training epoch.
For development purposes it's possible to run the script directly (without monai bundle calls)
python scripts/workflow.py --config_file configs/hyper_parameters.yaml ...
torchrun --nproc_per_node=gpu -m scripts/workflow.py --config_file configs/hyper_parameters.yaml ..Enable MLFlow logging by specifying "mlflow_tracking_uri" (can be local or remote URL).
python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mlflow_tracking_uri=http://127.0.0.1:8080Optionally use "--mlflow_run_name=.." to specify MLFlow experiment name, and "--mlflow_log_system_metrics=True/False" to enable logging of CPU/GPU resources (requires pip install psutil pynvml)
Test single GPU training:
python unit_tests/test_vista2d.py
Test multi-GPU training (may need to uncomment the "--standalone" in the unit_tests/utils.py file):
python unit_tests/test_vista2d_mgpu.py
Min GPU memory requirements 16Gb.
Vista2D codebase follows MONAI bundle format and its specifications. Make sure to run pre-commit before committing code changes to git
pip install pre-commit
python3 -m pre_commit run --all-filesJoin the conversation on Twitter @ProjectMONAI or join our Slack channel.
Ask and answer questions on MONAI VISTA's GitHub discussions tab.
The codebase is under Apache 2.0 Licence. The model weight is released under CC-BY-NC-SA-4.0. For various public data licenses please see data_license.txt.