--qos=course | For all course-related jobs | 72 | Course users only |
+
+Example:
+```bash
+#SBATCH --qos=course
+```
+
+## Course Directory
+
+Each student is assigned a dedicated **course directory**, and **all work must be done inside it**, including job scripts and outputs.
+
+Directory Format
+```bash
+/course/
+
+- ### [Open OnDemand on Wulver](6_intro_to_OnDemand.md)
+
+ ---
+
+ This webinar will introduce NJIT’s Open OnDemand portal, a browser-based gateway to the Wulver cluster and shared storage. With a focus on streamlining your HPC workflows, you will explore common scenarios and tasks through interactive demos. You will gain a detailed understanding of how to manage your files on the cluster, run interactive applications like Jupyter Notebook and RStudio, launch a full Linux desktop environment in your browser, and submit and monitor SLURM jobs. Additionally, you'll learn how to track resource usage and optimize your job performance for efficient computing on the Wulver cluster.
+
+ #### Key Highlights:
+ * Explore and manage your files on the cluster
+ * Run interactive tools like Jupyter Notebook and RStudio
+ * Launch a full Linux desktop environment in your browser
+ * Submit and monitor SLURM jobs
+ * Track resource usage and optimize job performance
+
+
+
+ [ Download Slides](../../assets/slides/Open_OnDemand_on_Wulver.pdf)
+
+
+
+
+
+- ### [Introduction to Linux](4_intro_to_linux.md)
+
+ ---
+
+ This is the fourth webinar of the 2025 Spring semester, introducing the basics of the Linux operating system. This session is designed to help new users become familiar with Linux, an essential skill for working in High-Performance Computing (HPC) environments.
+
+ #### Key Highlights:
+ * Basics of the Linux operating system
+ * Common commands and file system navigation
+ * Managing files, directories, and permissions
+ * Introduction to shell scripting for automation
+ * Connecting to remote systems and working with HPC cluster
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Linux.pdf)
+
+
+
+
+
+- ### [Python and Conda Environments in HPC: From Basics to Best Practices](3_conda_training.md)
+
+ ---
+
+ This is the third webinar of the 2025 Spring semester, focusing on an introductory understanding of using Python for HPC and effectively managing their Python environments using [Conda](conda.md)
+
+ #### Key Highlights:
+ * Learn how to manage Python environments using Conda.
+ * How to create Conda environments in different locations and install Python packages.
+ * Become familiar with common tools and libraries for scientific computing in Python.
+ * Import Conda environment to a different location.
+
+
+
+ [ Download Slides](../../assets/slides/Conda_training_Feb26.pdf)
+
+
+
+
+
+- ### [Introduction to Wulver: Accessing System & Running Jobs](2_intro_to_Wulver_II.md)
+
+ ---
+
+ This is the second webinar of the 2025 Spring semester, focusing on job submission, monitoring, and management on Wulver. This webinar also provides common tips for troubleshooting issues that users may encounter during job submission.
+
+ #### Key Highlights:
+ * How to Access HPC Software
+ * Introduction to SLURM and Its Role in HPC Environments
+ * Manage Slurm Jobs
+ * Troubleshooting Common Issues
+ * Slurm Interactive Jobs and Use GUI Apps
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Wulver_II_01_29_2025.pdf)
+
+
+
+
+
+- ### [Introduction to Wulver: Getting Started](1_intro_to_Wulver_I.md)
+
+ ---
+
+ This is the first webinar of the 2025 Spring semester introducing the NJIT HPC environment. This webinar provides basic information about our new High-Performance Computing (HPC) research cluster, [Wulver](wulver.md).
+
+ #### Key Highlights:
+ * Introduction to HPC (High Performance Computing)
+ * Hardware and architecture of Wulver
+ * Guidance on how to obtain an account and login to the cluster
+ * Data Storage systems
+ * Understanding allocations to utilize the shared resources
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Wulver_I_01_22_2025.pdf)
+
+
+
+## 2024 Fall
+
+
+- ### [Job Arrays and Advanced Submission Techniques for HPC](3_slurm_advanced.md)
+
+ ---
+
+ This is the final in a series of three webinars in the fall semester. designed to introduce researchers, scientists, and HPC users to the fundamentals of the containers. This session aims to provide useful information on submitting SLURM jobs efficiently by covering job arrays, job dependencies, checkpointing, and addressing common SLURM job issues.
+
+ #### Key Highlights:
+ * Understanding the concept and benefits of job arrays
+ * Syntax for submitting and managing job arrays
+ * Best practices for efficient array job design
+ * Dependency chains and complex workflows
+ * Resource optimization strategies
+ * Using SLURM's advanced options for improved job control
+ * Checkpointing the jobs and use of 3rd party checkpointing tool
+
+
+
+ [ Download Slides](../../assets/slides/HPC_Advanced_SLURM_11-20-2024.pdf)
+
+
+
+
+
+- ### [Introduction to Containers on Wulver](2_containers.md)
+
+ ---
+
+ This is the second in a series of three webinars in the fall semester, designed to introduce researchers, scientists, and HPC users to the fundamentals of the containers. Attendees will learn the fundamentals of Singularity, including installation, basic commands, and workflow, as well as how to create and build containers using definition files and existing Docker images. The training will cover executing containerized applications on HPC clusters and integrating with job schedulers like SLURM, while also addressing security considerations and performance optimization techniques.
+
+ #### Key Highlights:
+ * Introduction to containers and its role in HPC environments
+ * Fundamentals of Singularity, including installation, basic commands, and workflow
+ * Create and build containers using definition files and existing Docker images
+ * How to execute containerized applications on HPC clusters
+ * Use Containers via SLURM
+ * Performance optimization techniques
+
+
+
+
+ [ Download Slides](../../assets/slides/container_HPC_10-16-2024.pdf)
+
+
+
+
+
+- ### [SLURM Batch System Basics](1_slurm.md)
+
+ ---
+
+ This is the first in a series of three webinars in the fall semester. designed to introduce researchers, scientists, and HPC users to the fundamentals of the SLURM (Simple Linux Utility for Resource Management) workload manager. This virtual session will equip you with essential skills to effectively utilize HPC resources through SLURM.
+
+ #### Key Highlights:
+ * Introduction to SLURM and its role in HPC environments
+ * Basic SLURM commands for job submission, monitoring, and management
+ * How to write effective job scripts for various application types
+ * Understanding SLURM partitions, quality of service, and job priorities
+ * Best practices for resource requests and job optimization
+ * Troubleshooting common issues in job submission and execution
+
+
+
+
+ [ Download Slides](../../assets/slides/NJIT_HPC_Seminar-SLURM.pdf)
+
+
+
+## 2024 Spring
+ Since [Wulver](wulver.md) is quite different from the older cluster [Lochness](lochness.md), the HPC training programs are designed to guide both new and existing users on how to use the new cluster. The following trainings provide the basic information on
+
+ * Introduction to HPC
+ * Performance Optimization
+ * Job Submission and Management
+ * Managing Conda Environment
+
+If you still have any questions on HPC usage, please contact the [HPC Facilitator](contact.md).
+
+
+
+
+- ### Getting Started on Wulver: Session I
+
+ ---
+
+ This is the first in a series of three webinars introducing the NJIT HPC environment. This webinar provided the basic information in learning more about our new High Performance Computing (HPC) research cluster, [Wulver](wulver.md).
+
+ #### Key Highlights:
+ * HPC concepts
+ * Hardware and architecture of the Wulver cluster
+ * Guidance on how to obtain an account and receive an allocation to utilize the shared resources.
+
+
+
+ [ Download Slides](../../assets/slides/NJIT_HPC_Seminar-Part-I.pdf)
+
+
+
+
+- ### Getting Started on Wulver: Session II
+
+ ---
+
+ This session offered an overview of the environment on the Wulver cluster, including file management, working with the batch system (SLURM), and accessing software.
+ #### Key Highlights:
+ * HPC allocations
+ * Using [SLURM](../../Running_jobs/index.md)
+ * Job submissions
+
+
+
+ [ Download Slides](../../assets/slides/NJIT_HPC_Seminar-Part-II.pdf)
+
+
+- ### Introduction to Python and Conda
+
+ ---
+ Participants will gain an introductory understanding of using Python for HPC and effectively managing their Python environments using [Conda](conda.md). This knowledge will empower them to leverage the power of Python for their scientific computing needs on HPC systems.
+
+ #### Key Highlights:
+ * Learn how to manage Python environments for HPC using Conda.
+ * Become familiar with common tools and libraries for scientific computing in Python.
+
+
+
+ [ Download Slides](../../assets/slides/intro-to-Python-and-Conda.pdf)
+
+
+
+
diff --git a/docs/HPC_Events_and_Workshops/Workshop_and_Training_Videos/index.md b/docs/HPC_Events_and_Workshops/Workshop_and_Training_Videos/index.md
new file mode 100644
index 000000000..9f56cd2be
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/Workshop_and_Training_Videos/index.md
@@ -0,0 +1,116 @@
+# HPC Education and Training
+
+NJIT HPC provides practical training in high performance computing for students and researchers at various levels of expertise. HPC training for research professionals aims to enhance their capabilities in utilizing high-performance computing, data-intensive computing, and data analytics within their respective research fields.
+
+
+## 2025 Fall
+
+
+- ### [Intro to Wulver: Resources & HPC](../archived/index.md)
+
+ ---
+
+ This webinar provides essential information about the Wulver cluster, how to get an account, and allocation details, accessing installed software.
+
+ **Key Highlights:**
+
+ * Introduction to HPC (High Performance Computing)
+ * Hardware and architecture of Wulver
+ * Guidance on how to obtain an account and login to the cluster
+ * Data Storage systems
+ * Understanding allocations to utilize the shared resources
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Wulver_I_09_17_2025.pdf)
+
+
+
+
+
+- ### [Intro to Wulver: Job Scheduler & Submitting Jobs](../archived/index.md)
+
+ ---
+
+ This webinar provides the basic information on running jobs , how to run batch processing, and submit and manage the Slurm jobs.
+
+ **Key Highlights:**
+
+ * Access the software on Wulver
+ * Batch Processing
+ * Manage Slurm Jobs
+ * Troubleshooting Common Issues
+ * SlurmInteractive Jobs and Use GUI Apps
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Wulver_II_10_01_2025.pdf)
+
+
+
+
+
+- ### [Intro to Wulver: Focus on Job Efficiency](../archived/index.md)
+
+ ---
+
+ This webinar provides more in-depth features of SLURM, how to run dependency, array jobs to run efficiently on the cluster.
+
+ **Key Highlights:**
+
+ * Sbatch : Some Examples
+ * salloc command
+ * Job Dependencies
+ * Job Arrays
+ * Checkpointing
+
+
+
+ [ Download Slides](../../assets/slides/Intro_to_Wulver_III_10_08_2025.pdf)
+
+
+
+
+
+- ### [Conda for Shared Environments](../archived/index.md)
+
+ ---
+
+ This webinar provides an introductory understanding of using Python for HPC and effectively managing their Python environments.
+
+ **Key Highlights:**
+
+ * Access Python on Wulver
+ * Introduction to Conda environments
+ * Install, uninstall and upgrade packages
+ * Best Practices for managing conda environments
+ * Common Python libraries for scientific computing
+
+
+
+ [ Download Slides](../../assets/slides/conda_training_11-05-2025.pdf)
+
+
+
+
+
+
+- ### [HPC User Meeting - Introduction to MIG](../archived/index.md)
+
+ ---
+
+ This in-person and virtual session provide an introductory understanding of using Multi-Instance GPUs (MIGs) on Wulver.
+
+ **Key Highlights:**
+
+ * What is MIG?
+ * Why MIG on Wulver?
+ * MIG Configuration Example
+ * Submitting Jobs (`srun` & `sbatch`)
+ * New Billing Model
+
+
+
+ [ Download Slides](../../assets/slides/Wulver_MIG_Dec03_2025.pdf)
+
+
\ No newline at end of file
diff --git a/docs/HPC_Events_and_Workshops/archived/2024/1_slurm.md b/docs/HPC_Events_and_Workshops/archived/2024/1_slurm.md
new file mode 100644
index 000000000..1575ff7a6
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/archived/2024/1_slurm.md
@@ -0,0 +1,29 @@
+---
+hide:
+ - toc
+---
+
+
+
+# SLURM Batch System Basics
+
+Join us for an informative webinar designed to introduce researchers, scientists, and HPC users to the fundamentals of the SLURM (Simple Linux Utility for Resource Management) workload manager. This virtual session will equip you with essential skills to effectively utilize HPC resources through SLURM.
+
+- Date: Sep 18th 2024
+- Location: Virtual
+- Time: 2:30 PM - 3:30 PM
+
+## Topics Covered
+
+* Introduction to SLURM and its role in HPC environments
+* Basic SLURM commands for job submission, monitoring, and management
+* How to write effective job scripts for various application types
+* Understanding SLURM partitions, quality of service, and job priorities
+* Best practices for resource requests and job optimization
+* Troubleshooting common issues in job submission and execution
+
+Our experienced HPC specialists will guide you through practical examples and provide tips for efficient use of SLURM in your research workflows. Whether you're new to HPC or looking to refine your SLURM skills, this webinar will help you maximize your productivity on SLURM-based clusters.
+
+## Registration
+
+Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#slurm-batch-system-basics) for the webinar recording and slides.
diff --git a/docs/HPC_Events_and_Workshops/archived/2024/2_containers.md b/docs/HPC_Events_and_Workshops/archived/2024/2_containers.md
new file mode 100644
index 000000000..f8ec42165
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/archived/2024/2_containers.md
@@ -0,0 +1,29 @@
+---
+hide:
+ - toc
+---
+
+
+
+# Introduction to Containers on Wulver
+
+The HPC training event on using Singularity containers provides participants with a comprehensive introduction to container technology and its advantages in high-performance computing environments.
+
+- Date: Oct 16th 2024
+- Location: Virtual
+- Time: 2:30 PM - 3:30 PM
+
+## Topics Covered
+
+* Introduction to containers and its role in HPC environments
+* Fundamentals of Singularity, including installation, basic commands, and workflow
+* Create and build containers using definition files and existing Docker images
+* How to execute containerized applications on HPC clusters
+* Use Containers via SLURM
+* Performance optimization techniques
+
+
+## Registration
+
+Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#introduction-to-containers-on-wulver) for the webinar recording and slides.
+
diff --git a/docs/HPC_Events_and_Workshops/archived/2024/3_slurm_advanced.md b/docs/HPC_Events_and_Workshops/archived/2024/3_slurm_advanced.md
new file mode 100644
index 000000000..b5be8a53a
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/archived/2024/3_slurm_advanced.md
@@ -0,0 +1,32 @@
+---
+hide:
+ - toc
+---
+
+
+
+# Job Arrays and Advanced Submission Techniques for HPC
+
+Elevate your High-Performance Computing skills with our advanced SLURM webinar! This session is designed for HPC users who are familiar with basic SLURM commands and are ready to dive into more sophisticated job management techniques.
+
+- Date: Nov 20th 2024
+- Location: Virtual
+- Time: 2:30 PM - 3:30 PM
+
+## Topics Covered
+
+### Job Arrays
+* Understanding the concept and benefits of job arrays
+* Syntax for submitting and managing job arrays
+* Best practices for efficient array job design
+
+### Advanced Job Submission Techniques
+* Dependency chains and complex workflows
+* Resource optimization strategies
+* Using SLURM's advanced options for improved job control
+* Techniques for balancing resource requests and job efficiency
+
+## Registration
+
+Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#job-arrays-and-advanced-submission-techniques-for-hpc) for the webinar recording and slides.
+
diff --git a/docs/HPC_Events_and_Workshops/archived/2024/4_nvidia.md b/docs/HPC_Events_and_Workshops/archived/2024/4_nvidia.md
new file mode 100644
index 000000000..1cf059c85
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/archived/2024/4_nvidia.md
@@ -0,0 +1,33 @@
+---
+hide:
+ - toc
+---
+
+# NVIDIA Workshop
+
+## Fundamentals of Accelerated Data Science
+
+Learn to use GPU-accelerated resources to analyze data. This is an intermediate level workshop that is intended for those who have some familiarity with Python, especially NumPy and SciPy libraries.
+
+- Date: July 15th 2024
+- Location: GITC 3700
+- Time: 9 AM - 5 PM
+
+### Schedule
+
+| Time | Topic |
+|:----:|:-----:|
+| 9:00 AM - 9:15 AM | Introduction |
+| 9:15 AM - 11:30 AM | GPU-Accelerated Data Manipulation |
+| 11:30 AM - 12:00 PM | GPU-Accelerated Machine Learning |
+| 12:00 PM - 1:00PM | Lunch |
+| 1:00 PM - 2:30 PM | GPU-Accelerated Machine Learning (contd) |
+| 2:45 PM - 4:45 PM | Project: Data Analysis to Save the UK |
+| 4:45 PM - 5:00 PM | Assessment and Q&A |
+
+Coffee and Lunch will be provided. See more detail about the workshop [here](https://www.nvidia.com/content/dam/en-zz/Solutions/deep-learning/deep-learning-education/DLI-Workshop-Fundamentals-of-Accelerated-Data-Science-with-RAPIDS.pdf).
+
+
+
+Registration is now closed.
+
diff --git a/docs/HPC_Events_and_Workshops/archived/2024/5_symposium.md b/docs/HPC_Events_and_Workshops/archived/2024/5_symposium.md
new file mode 100644
index 000000000..f2dcfe34a
--- /dev/null
+++ b/docs/HPC_Events_and_Workshops/archived/2024/5_symposium.md
@@ -0,0 +1,71 @@
+---
+hide:
+ - toc
+title: HPC Research Symposium
+---
+
+
+
+# HPC Research Symposium
+
+This past year has been transformative for HPC research at NJIT. The introduction of our new shared HPC cluster, Wulver, has expanded our computational capacity and made research into vital areas more accessible to our faculty. The Advanced Research Computing Services group of Information Services and Technology, in collaboration with Dell Technologies, invites you to a symposium on July 16, 2024, to celebrate the launch of Wulver.
+
+The Symposium will feature a keynote from Anthony Dina, Global Field CTO for Unstructured Data Solutions at Dell Technologies, and invited speakers from NJIT, Dibakar Datta from the Department of Mechanical and Industrial Engineering, and Cambridge Computer Services, Jose Alvarez. The Symposium will also feature several lightning talks from NJIT researchers highlighting the use of High Performance Computing resources in their research.
+
+Please join us to learn how our researchers use HPC resources and connect with the NJIT HPC community.
+
+
+- Date: July 16th, 2024
+- Location: Campus Center Atrium
+- Time: 9AM - 5PM
+
+## Agenda
+
+| Time | Session | Title
+|:-----:|:------:|:-----:
+| 9:00 - 9:15 | Welcome Remarks | **Ed Wozencroft**, Vice President for Digital Strategy & CIO |
+| 9:15 - 9:35 | Research Computing @ NJIT | _Overview of Research Computing Services_ **Gedaliah Wolosh**, Director High Performance Research Computing | +| 9:40 - 10:30 | Keynote | _It’s Time Research Behaves More Like Formula 1_
**Anthony Dina**, Global Field CTO for the Unstructured Data Solutions at Dell Technologies | +| 10:30 - 10:40 | Break | +| 10:40 - 11:20 | Invited Speaker | _Electro-Chemo-Mechanics of Multiscale Active Materials for Next-Generation Energy Storage_
**Dibakar Datta**, Associate Professor, Mechanical & Industrial Engineering | +| 11:20 - 12:00 | Lightning Talks I | _Hemodynamics and Cancer Cell Transport in a Tortuous in vivo Microvessel_
**Ali Kazempour**, Peter Balogh Research Group, Mechanical & Industrial Engineering | +| | | _Running Two-phase Flows on Wulver: Introduction to Basilisk_
**Matthew Cho**, Shahriar Afkhami Research Group, Mathematical Sciences +| | | _Temporal Super-Resolution of Solar Images with Generative AI_
**Jialing Li**, Jason Wang Research Group, Computer Science | +| | | _Numerical study of Thermo-Marangoni flow induced by a warm plate_
**Shivam Verma**, Pushpendra Singh Research Group, Mechanical & Industrial Engineering | +| 12:10 - 13:00 | Lunch | +| 13:00 - 13:30 | Invited Speaker | _Introduction to Grace Hopper and ARM Technology in Higher Education_
**Jose Alvarez**, Vice President Research Computing HPC/AI, Cambridge Computer Services | +| 13:30- 14:20 | Lightning Talks II | _Inference of Nullability Annotations using Machine Learning_
**Kazi Amanul Islam Siddiqui**, Martin Kellogg Research Group, Computer Science | +| | | _Deep Learning for Spatial Image Super-Resolution of Solar Observations_
**Chunhui Xu**, Jason Wang Research Group, Computer Science | +| | | _Volume Integral Method for Electromagnetic Equations_
**Matthew Cassini**, Thi Phong Nguyen Research Group, Mathematical Sciences | +| | | _Enhancing Region-based Image Captioning with Contextual Feature Exploitation_
**Al Shahriar Rubel**, Fadi Deek Research Group, Informatics | +| | | _Instability between the two-layer Poiseuille flow with the VOF method_
**Nastaran Rezaei**, Shahriar Afkhami Research Group, Mathematical Sciences | +| 14:20 - 14:30 | Break | | +| 14:30 - 15:00 | Research Computing @ NJIT | _Introducing Open OnDemand web portal: New Technologies on Wulver_
**Kate Cahill**, Associate Director High Performance Research Computing | +| 15:00 - 15:50 | Lightning Talks III | _Red Blood Cell Modeling Reveals 3D Angiogenic Wall Shear Stress Patterns_
**Mir Md Nasim Hossain**, Peter Balogh Research Group, Mechanical & Industrial Engineering | +| | | _Understanding and Forecasting Space Weather with Artificial Intelligence_
**Hongyang Zhang**, Jason Wang Research Group, Computer Science| +| | | _Entropic pressure on fluctuating solid membranes_
**Rubayet Hassan**, Fatemeh Ahmadpoor Research Group, Mechanical & Industrial Engineering | +| | | _Investigation of PFAS Adsorption on Functionalized COF-300_
**Daniel Mottern**, Joshua Young Research Group, Chemical & Materials Engineering | +| | | _Large Language models (LLM) for hardware_
**Deepak Vungarala**, Shaahin Angizi Research Group, Electrical and Computer Engineering | +| 15:50 - 16:00 | Break | +| 16:00 - 17:00 | Poster Session | _Molecular Dynamic study on Ar nano-bubble stability in water_
**Targol Teymourian**, Jay Meegoda Research Group, Civil & Environmental Engineering | +| | | _Possible mechanism for sonolytic degradation of PFAS_
**Laura Nwanebu**, Jay Meegoda Research Group, Civil & Environmental Engineering | +| | | _Entropic Force Near Fluctuating Surface_
**Rubayet Hassan**, Fatemeh Ahmadpoor Research Group, Mechanical & Industrial Engineering | +| | | _DFT investigations of the enantioselective phase-transfer-catalyzed aza-Michael cyclization of ureas_
**Diana Marlen Castaneda Bagatella**, Pier Champagne Research Group, Chemistry and Environmental Science | +| | | + + diff --git a/docs/HPC_Events_and_Workshops/archived/2024/6_slurm_workshop.md b/docs/HPC_Events_and_Workshops/archived/2024/6_slurm_workshop.md new file mode 100644 index 000000000..85ffba5dc --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2024/6_slurm_workshop.md @@ -0,0 +1,12 @@ +--- +hide: + - toc +--- + +# SLURM Workload Manager Workshop + +Advanced Research Computing Services in collaboration with SchedMD is pleased to announce a two-day workshop on SLURM Workload Manager on **August 13-14, 2024**. This immersive 2-day experience will take you through comprehensive technical scenarios with lectures, demos, and workshop lab environments. The Slurm trainer will assist in identifying commonalities between previously used resources and schedulers, offering increased understanding and adoption of [SLURM](slurm.md) job scheduling, resource management, and troubleshooting techniques. + +Registration is now closed. + + diff --git a/docs/HPC_Events_and_Workshops/archived/2024/7_Intro_to_MPI_Workshop.md b/docs/HPC_Events_and_Workshops/archived/2024/7_Intro_to_MPI_Workshop.md new file mode 100644 index 000000000..35209f771 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2024/7_Intro_to_MPI_Workshop.md @@ -0,0 +1,12 @@ +--- +hide: + - toc +--- + +# Intro to MPI Workshop + +NJIT is an in-person, satellite location for a two-day HPC workshop hosted by the **Pittsburgh Supercomputing Center (PSC)** on December 10 & 11. This is a great introduction to using MPI programming to scale up your computational research. Attendees will leave with a working knowledge of how to write scalable codes using MPI – the standard programming tool of scalable parallel computing. + +Registration is now closed. + +To download the training materials, check [MPI Workshop Agenda](https://t.e2ma.net/click/mhwgmh/mtd2z9gb/a459ow). diff --git a/docs/HPC_Events_and_Workshops/archived/2025/1_intro_to_Wulver_I.md b/docs/HPC_Events_and_Workshops/archived/2025/1_intro_to_Wulver_I.md new file mode 100644 index 000000000..af60dbc89 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/1_intro_to_Wulver_I.md @@ -0,0 +1,25 @@ +--- +hide: + - toc +--- + +# Introduction to Wulver: Getting Started + +This is the first webinar of the 2025 Spring semester introducing the NJIT HPC environment. This webinar provides basic information about our new High-Performance Computing (HPC) research cluster, [Wulver](wulver.md). + +- Date: Jan 22nd 2025 +- Location: Virtual +- Time: 2:30 PM - 3:30 PM + +## Topics Covered + +* Introduction to HPC (High Performance Computing) +* Hardware and architecture of Wulver +* Guidance on how to obtain an account and login to the cluster +* Understanding allocations to utilize the shared resources + +Our experienced HPC specialists will guide you through practical examples and provide tips for proper use of cluster in your research workflows. + +## Registration + +Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#introduction-to-wulver-getting-started) for the webinar recording and slides. diff --git a/docs/HPC_Events_and_Workshops/archived/2025/2_intro_to_Wulver_II.md b/docs/HPC_Events_and_Workshops/archived/2025/2_intro_to_Wulver_II.md new file mode 100644 index 000000000..07a36dd19 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/2_intro_to_Wulver_II.md @@ -0,0 +1,24 @@ +--- +hide: + - toc +--- + +# Introduction to Wulver: Accessing System & Running Jobs + +This is the second webinar of the 2025 Spring semester introducing the NJIT HPC environment. This webinar provided the basic information in learning more about our new High Performance Computing (HPC) research cluster, [Wulver](wulver.md). + +- Date: Jan 29th 2025 +- Location: Virtual +- Time: 2:30 PM - 3:30 PM + +## Topics Covered + +* HPC allocations +* How to access HPC software +* Introduction to SLURM and its role in HPC environments +* Basic SLURM commands for job submission, monitoring, and management +* Troubleshooting common issues in job submission and execution + +## Registration + +Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#introduction-to-wulver-accessing-system-running-jobs) for the webinar recording and slides. diff --git a/docs/HPC_Events_and_Workshops/archived/2025/3_conda_training.md b/docs/HPC_Events_and_Workshops/archived/2025/3_conda_training.md new file mode 100644 index 000000000..1d4ab8eff --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/3_conda_training.md @@ -0,0 +1,22 @@ +--- +hide: + - toc +--- + +# Python and Conda Environments in HPC: From Basics to Best Practices + +This is the third webinar of the 2025 Spring semester introducing the NJIT HPC environment. Participants will gain an introductory understanding of using Python for HPC and effectively managing their Python environments using [Conda](conda.md). This knowledge will empower them to leverage the power of Python for their scientific computing needs on HPC systems. + +- Date: Feb 26th 2025 +- Location: Virtual +- Time: 2:30 PM - 3:30 PM + +## Topics Covered + +* Learn how to manage Python environments using Conda. +* How to create Conda environments in different locations and install Python packages. +* Become familiar with common tools and libraries for scientific computing in Python. + + +## Registration +Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#python-and-conda-environments-in-hpc-from-basics-to-best-practices) for the webinar recording and slides. \ No newline at end of file diff --git a/docs/HPC_Events_and_Workshops/archived/2025/4_intro_to_linux.md b/docs/HPC_Events_and_Workshops/archived/2025/4_intro_to_linux.md new file mode 100644 index 000000000..bd16da433 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/4_intro_to_linux.md @@ -0,0 +1,24 @@ +--- +hide: + - toc +--- + +# Introduction to Linux + +This is the fourth webinar of the 2025 Spring semester, introducing the basics of the Linux operating system. This session is designed to help new users become familiar with Linux, an essential skill for working in High-Performance Computing (HPC) environments. + +- Date: March 26th 2025 +- Location: Virtual +- Time: 2:30 PM - 3:30 PM + +## Topics Covered + +* Basics of the Linux operating system +* Common commands and file system navigation +* Managing files, directories, and permissions +* Introduction to shell scripting for automation +* Connecting to remote systems and working with HPC cluster + + +## Registration +Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#introduction-to-linux) for the webinar recording and slides. diff --git a/docs/HPC_Events_and_Workshops/archived/2025/5_parallel_computing_with_matlab.md b/docs/HPC_Events_and_Workshops/archived/2025/5_parallel_computing_with_matlab.md new file mode 100644 index 000000000..ce1fcd768 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/5_parallel_computing_with_matlab.md @@ -0,0 +1,26 @@ +--- +hide: + - toc +--- + +# Parallel Computing with MATLAB: Hands on workshop + +During this hands-on workshop, we will introduce parallel and distributed computing in MATLAB with a focus on speeding up application codes and offloading computers. By working through common scenarios and workflows using hands-on demos, you will gain a detailed understanding of the parallel constructs in MATLAB, their capabilities, and some of the common hurdles that you'll encounter when using them. + +- Date: April 16th 2025 +- Location: Virtual +- Time: 1:00 PM - 4:00 PM +- Hosted by Mathworks - A MATLAB account is required to participate.. + +## Topics Covered + +* Multithreading vs multiprocessing +* When to use parfor vs parfeval constructs +* Creating data queues for data transfer +* Leveraging NVIDIA GPUs +* Parallelizing Simulink models +* Working with large data + + +## Registration +Registration is now closed. diff --git a/docs/HPC_Events_and_Workshops/archived/2025/6_intro_to_OnDemand.md b/docs/HPC_Events_and_Workshops/archived/2025/6_intro_to_OnDemand.md new file mode 100644 index 000000000..c3d815161 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/6_intro_to_OnDemand.md @@ -0,0 +1,25 @@ +--- +hide: + - toc +--- + +# Open OnDemand on Wulver + +[Open OnDemand](https://ondemand.njit.edu) is a browser-based gateway to NJIT's Wulver cluster and shared storage. + +- Date: April 30th 2025 +- Location: Virtual +- Time: 2:30 PM - 3:30 PM + +## Topics Covered + +* Explore and manage your files on the cluster +* Run interactive tools like Jupyter Notebook and RStudio +* Launch a full Linux desktop environment in your browser +* Submit and monitor SLURM jobs +* Track resource usage and optimize job performance + + +## Registration +Registration is now closed. Check the [HPC training](../../Workshop_and_Training_Videos/index.md#open-ondemand-on-wulver) for the webinar recording and slides. + diff --git a/docs/HPC_Events_and_Workshops/archived/2025/7_MATLAB_on_Wulver.md b/docs/HPC_Events_and_Workshops/archived/2025/7_MATLAB_on_Wulver.md new file mode 100644 index 000000000..4ec74f46c --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/7_MATLAB_on_Wulver.md @@ -0,0 +1,36 @@ +--- +hide: + - toc +--- + +# HPC Summer Workshop: MATLAB Parallel Computing Hands-On Using Wulver + +## Overview +Join us for an interactive webinar hosted at NJIT's HPC facility, where MATLAB expert (Evan Cosgrov) +will guide participants through practical techniques for accelerating code and workflows using MATLAB’s parallel computing tools. +Through live demonstrations and guided examples, you’ll gain a solid understanding of how to parallelize MATLAB code, overcome common challenges, and optimize performance across distributed computing environments. + +Each participant will have access to the [OnDemand Matlab server](https://ondemand.njit.edu/pun/sys/dashboard/batch_connect/sys/jupyter-matlab/session_contexts/new) running on [Wulver](https://hpc.njit.edu/). + +## Guide to access Matlab via OnDemand: +Users need use `Jupyter Matlab Proxy` to use MATLAB on OnDemand. + +## Date and Location: +- Date: June 12th 2025 +- Location: Campus Center 235 +- Time: 1:00 PM - 4.00 PM + +## Topics Covered + +* How to identify bottlenecks in serial MATLAB code and convert them to run in parallel. +* Practical differences between `parfor` and `parfeval`, and how to choose the right one. +* Creating asynchronous tasks and managing outputs with DataQueue. +* Running Simulink models in parallel and accelerating simulation tasks. +* Leveraging GPU resources to boost performance in compute-intensive operations. +* Working with large datasets using tall arrays and distributed arrays. +* Best practices for launching and managing MATLAB parallel pools. + + +## Registration +Registration is now closed. + diff --git a/docs/HPC_Events_and_Workshops/archived/2025/8_PSC_Machine_Learning_workshop.md b/docs/HPC_Events_and_Workshops/archived/2025/8_PSC_Machine_Learning_workshop.md new file mode 100644 index 000000000..9067d77ba --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/2025/8_PSC_Machine_Learning_workshop.md @@ -0,0 +1,22 @@ +--- +hide: + - toc +--- + +# HPC Summer Workshop: PSC Machine Learning and BIG DATA Workshop + +## Overview +The Pittsburgh Supercomputing Center is pleased to present a Machine Learning and Big Data workshop. This workshop will focus on topics including big data analytics and machine learning with Spark, and deep learning using Tensorflow. This will be an IN PERSON event hosted by various satellite sites, there WILL NOT be a direct to desktop option for this event. + + +## Date and Location: +- Date: July 29-30, 2025 +- Location: Room 2315A GITC +- Time: 11:00 AM - 5.00 PM + +## Topics Covered +Check details at [PSC Machine Learning and BIG DATA Workshop](https://support.access-ci.org/events/8089) + +## Registration +Registration is now closed. + diff --git a/docs/HPC_Events_and_Workshops/archived/index.md b/docs/HPC_Events_and_Workshops/archived/index.md new file mode 100644 index 000000000..11c29b91b --- /dev/null +++ b/docs/HPC_Events_and_Workshops/archived/index.md @@ -0,0 +1,115 @@ +--- +hide: + - toc +--- + +# Archived HPC Workshops + +## 2025 + + +=== "Fall" + + ```python exec="on" + import re + import pandas as pd + + df = pd.read_csv('docs/assets/tables/trainings/2025_fall.csv', keep_default_na=False) + + def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + + df = df.applymap(fix_cell) + print(df.to_markdown(index=False)) + ``` + +=== "Summer" + + ```python exec="on" + import re + import pandas as pd + + df = pd.read_csv('docs/assets/tables/trainings/2025_summer.csv', keep_default_na=False) + + def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + + df = df.applymap(fix_cell) + print(df.to_markdown(index=False)) + ``` +=== "Spring" + + ```python exec="on" + import re + import pandas as pd + + df = pd.read_csv('docs/assets/tables/trainings/2025_spring.csv', keep_default_na=False) + + def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + + df = df.applymap(fix_cell) + print(df.to_markdown(index=False)) + ``` + +## 2024 + +=== "Fall" + + ```python exec="on" + import re + import pandas as pd + + df = pd.read_csv('docs/assets/tables/trainings/2024_fall.csv', keep_default_na=False) + + def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + + df = df.applymap(fix_cell) + print(df.to_markdown(index=False)) + ``` + +=== "Summer" + + ```python exec="on" + import re + import pandas as pd + + df = pd.read_csv('docs/assets/tables/trainings/2024_summer.csv', keep_default_na=False) + + def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + + df = df.applymap(fix_cell) + print(df.to_markdown(index=False)) + ``` diff --git a/docs/HPC_Events_and_Workshops/index.md b/docs/HPC_Events_and_Workshops/index.md new file mode 100644 index 000000000..780c030e7 --- /dev/null +++ b/docs/HPC_Events_and_Workshops/index.md @@ -0,0 +1,33 @@ +--- +hide: + - toc +--- + +# HPC Events + +## 2026 Spring +Please check our workshop schedule for this spring season. Expand each section to view more details about the event. For webinars, the links will be sent to your email once you register. The links to slides and recordings will be updated after each webinar. For the HPC User Meeting, users are encouraged to register using the form provided in the registration link; however, registration is not mandatory. If you forget or miss registering, you are still welcome to stop by the location listed in the schedule below. + +```python exec="on" +import re +import pandas as pd + +df = pd.read_csv('docs/assets/tables/trainings/2026_spring.csv', keep_default_na=False) + +def fix_cell(s): + if not isinstance(s, str): + return s + s = re.sub(r'(\()([^)]*?)index\.md', r'\1\2', s) + s = re.sub(r'(\b6_[\w\-.]+)\.md\b', r'\1', s) + s = s.replace('(//', '(/') + + return s + +df = df.applymap(fix_cell) +print(df.to_markdown(index=False)) +``` +!!! info "Archived Workshops" + + Click here to review our [past workshops](archived/index.md)! + + diff --git a/docs/MIG/index.md b/docs/MIG/index.md new file mode 100644 index 000000000..53e1db287 --- /dev/null +++ b/docs/MIG/index.md @@ -0,0 +1,76 @@ +# MIG Overview + +MIG (Multi-Instance GPU) is a technology introduced by NVIDIA starting with its Ampere architecture (e.g., A100). It enables a single physical GPU to be partitioned into multiple smaller, isolated GPU instances — each with dedicated compute cores, memory, cache, and bandwidth. These instances function independently and appear to software as discrete GPUs. + +This allows multiple users or processes to simultaneously run GPU workloads without interfering with one another, improving resource utilization, reducing wait times in job queues, and increasing throughput in shared computing environments like Wulver. + + +## Why MIG on Wulver + + +
+  +
+  +
+
+
+The plots above show cluster-wide usage of the A100 GPUs over the last 3 months. In short: GPUs were **heavily allocated** but **lightly utilized**, which means lots of capacity sat idle behind single jobs.
+
+- **Allocated vs. used**: GPUs were reserved ~85–95% of the time, while average compute utilization was ~25–50% and memory utilization stayed mostly under 20%.
+
+- **Impact**: Long queue times and poor overall throughput when full GPUs are booked for workloads that only need a fraction of the device.
+
+- **MIG rationale**: Partitioning A100s into 10/20/40 GB slices lets multiple right-sized jobs run concurrently, improving SU efficiency and time-to-results without requiring more hardware.
+
+## MIG Implementation in Wulver
+
+MIG is implemented on selected **NVIDIA A100 80GB** GPUs. Wulver currently supports the following MIG configurations:
+
+- 10gb
+- 20gb
+- 40gb
+
+These profiles correspond to different partitions of compute and memory resources from the A100 80GB GPU. You can view a full comparison in the [Profile Comparison section](../MIG/profile-comparison.md).
+
+Each profile maps to a Service Unit (SU) usage factor that reflects its computational weight — ranging from 2 SU/hour for a 10gb instance up to 16 SU/hour for a full GPU. You can check full SU overview of MIG [here](../MIG/job-submission-and-su-charges.md#understanding-su-charges).
+
+MIGs address key challenges in shared environments:
+
+- **Fair resource sharing**: MIG enables multiple users to share a single GPU without stepping on each other’s performance.
+
+- **Right-sizing workloads**: Users can request GPU capacity that matches their actual workload requirements.
+
+- **Improved scheduling efficiency**: MIG instances are smaller and easier to schedule than full GPUs.
+
+- **Reduced idle time**: MIG reduces GPU underutilization by splitting large resources into usable chunks.
+
+
+## Why You Should Use MIG Instances
+
+**Lower SU Cost for Smaller Workloads**: +
+  +
++MIG lets you select a GPU slice that meets your job’s needs without paying for more power than you use. +For example: + +- A small training job or inference script may only require `10–20` GB of GPU memory. +- Running such a job on a `10gb` or `20gb` MIG instance will consume only a fraction of the SUs compared to using a full GPU. + +**No Need to Reserve a Full GPU**:
+Some jobs — like unit testing, data preprocessing with GPU acceleration, or light model inference — simply don’t need the full resources of an 80GB A100. MIG allows users to avoid bottlenecks and free up resources by choosing an appropriately sized instance.This is especially valuable when: + +- Cluster demand is high +- Your jobs don’t need massive memory or compute +- You're trying to run multiple parallel tasks independently + +This is especially important if you’re working within a research group’s annual SU allocation on Wulver. + +**Isolated and Predictable Performance**:
+Each MIG instance has dedicated resources — memory, L2 cache, compute cores — and is logically isolated from others on the same GPU. That means: + +- No performance interference from noisy neighbors +- Consistent and reliable job behavior +- Easier debugging and performance tuning + +MIG is compatible with CUDA, cuDNN, PyTorch, TensorFlow, and most GPU-accelerated libraries — no code changes are typically required. + +!!! info + + MIG is not implemented on all GPUs in Wulver. \ No newline at end of file diff --git a/docs/MIG/job-submission-and-su-charges.md b/docs/MIG/job-submission-and-su-charges.md new file mode 100644 index 000000000..f9b57228e --- /dev/null +++ b/docs/MIG/job-submission-and-su-charges.md @@ -0,0 +1,85 @@ +# MIG Job Submission and SU Charges + +When submitting jobs on Wulver's MIG-enabled A100 GPUs, you must explicitly request the desired MIG profile using the `--gres` directive in your SLURM script. + +| GPU MIG | Slurm Directive | +|---------|:----------------------------------:| +| 10G MIG | `--gres=gpu:a100_10g:1 ` | +| 20G MIG | `--gres=gpu:a100_20g:1 ` | +| 40G MIG | `--gres=gpu:a100_40g:1 ` | + + +!!! note + If you want to see a job script example of requesting a full GPU, please refer to the sample [GPU job scripts](../Running_jobs/batch-jobs.md/#example-of-batch-job-slurm-script-gpu-nodes). + +## Running Jobs with MIG + +=== "Sample SLURM Script for a MIG Job" + + ```shell + #!/bin/bash -l + #SBATCH --job-name=gpu_job + #SBATCH --output=%x.%j.out + #SBATCH --error=%x.%j.err + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace with PI's UCID + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --gres=gpu:a100_10g:1 # Change to 20g or 40g as needed + #SBATCH --time=59:00 + #SBATCH --mem-per-cpu=4000M + + srun ./myexe + ``` + +=== "Interactive session with MIG" + + ```shell + $srun --partition=gpu \ + --account=$PI_ucid \ + --qos=standard \ + --gres=gpu:a100_10g:1 \ + --time=00:59:00 \ + --pty bash + ``` + +!!!warning + You cannot run your job using multiple MIG instances. For example, `--gres=gpu:a100_10g:2` will allocate two instances of the 10G MIG, but it will either raise an error or some jobs may assume it as a single MIG, even if multiple instances are requested. + +## Understanding SU Charges + +Wulver uses a Service Unit (SU) model to track computing usage. Your job's SU cost is based on: + +- CPU usage +- Memory request +- GPU memory allocation (via MIG) + +Each component contributes to the SU calculation. The SU cost is charged per hour using the formula: + +``` +SU = MAX(#CPUs, Memory (in GB) / 4) + 16 × (GPU memory requested / 80GB) +``` + +!!! info + GPU memory requested is based on the MIG profile, not your actual memory usage during the job. + + +| SLURM Directive | SU | Explaination | +|---------------------|:---------:|:---------:| +| 4 CPUs + 10MIG | MAX(4, 4*4G/4G) + 16 * (10G/80G) = 6 | Since no memory requireemnt is specified, SU is charged based on the same number of CPUs and 10G of GPU memory | +| 4 CPUs + 20MIG | MAX(4, 4*4G/4G) + 16 * (20G/80G) = 8 | SU is charged based on the same number of CPUs and 20G of GPU memory | +| 4 CPUs + 40MIG | MAX(4, 4*4G/4G) + 16 * (40G/80G) = 12 | SU is charged based on the same number of CPUs and 40G of GPU memory | +| 4 CPUs + Full GPU | MAX(4, 4*4G/4G) + 16 * (80G/80G) = 20 | SU is charged based on the same number of CPUs and 80G of GPU (A full GPU) memory | +| 4 CPUs + `--mem=64G` + Full GPU | MAX(4, 64G/4G) + 16 * (80G/80G) = 32 | The MAX function the evaluates the maximum of 4 SUs (from CPUs), and 64G/4G= 16 SUs (from memory). In addition, 16 SUs are charged from 80G of GPU (A full GPU) memory, bringing the total SU charge to 32 SUs | +| 4 CPUs + `--mem-per-cpu=8G` + Full GPU | MAX(4, 4*8G/4G) + 16 * (80G/80G) = 24 | The MAX function the evaluates the maximum of 4 SUs (from CPUs), and 4*8G/4G= 8 SUs (from memory). In addition, 16 SUs are charged from 80G of GPU (A full GPU) memory, bringing the total SU charge to 24 SUs | + + +## Tips for Efficient Job Submission ***(Think Fit, Not Power)*** + +- Choose the profile that fits your workload, not the biggest one available. You’ll save SUs, get scheduled faster, and help the cluster stay responsive for everyone. +- Avoid requesting a full GPU unless your job cannot run on a MIG. +- Combine small jobs using job arrays or batching when possible. +- Need help estimating SUs? Try submitting test jobs with `--time=10:00` and reviewing the actual SU usage via the job summary. +- By default, each CPU is allocated `4 GB` of memory. If your job requires more, you can request additional memory using `--mem`. Please ensure you request only as much as your job needs, as over-allocating memory will increase your SU usage. Refer to the formula above for details. +- MIG is designed to make high-performance GPUs accessible and efficient — take advantage of it wisely. diff --git a/docs/MIG/performance_testing.md b/docs/MIG/performance_testing.md new file mode 100644 index 000000000..96305836b --- /dev/null +++ b/docs/MIG/performance_testing.md @@ -0,0 +1,71 @@ +# Performance Testing Overview + +To help users select the appropriate MIG profile for their workloads, we conducted benchmark tests using LLM fine-tuning, PyTorch training, and GROMACS molecular dynamics simulations. Tests were run on the NVIDIA A100 GPUs (full 80 GB and MIG profiles) available on Wulver. + +The results below show differences in runtime, accuracy, memory usage, and service unit (SU) cost across profiles. Observations and notes are included to explain results. + +## List of Benchmark Tests +=== "GROMACS" + + The GROMACS benchmark test was conducted for different MIG profiles and a full GPU. The results suggest that the 40G MIG shows better performance compared to a full 80G GPU. We also calculated the cost/performance parameter, which is obtained by taking the ratio of SU consumption to performance. Based on this parameter, the 20G MIG is the best choice. However, if users prefer performance over cost/performance, then the 40G MIG would be the recommended option. + + ```python exec="on" + import pandas as pd + import numpy as np + df = pd.read_csv('docs/assets/tables/MIG/gromacs_performance.csv') + df.replace(np.nan, 'NA', inplace=True) + print(df.to_markdown(index=False)) + ``` + +=== "LLM Fine-Tuning" + + The benchmark script fine-tunes the Qwen 1.5B Instruct model on the Alpaca-cleaned dataset using QLoRA. Training is done with 4-bit quantization to save memory and LoRA adapters so that only a small set of parameters are updated. The Hugging Face TRL `SFTTrainer` handles training, while the script also logs runtime, GPU/CPU memory, and tokens processed per second. The setup runs consistently on both full NVIDIA's A100 80GB GPU and different MIG slices (10 GB, 20 GB, 40 GB), making it useful for comparing speed and cost across profiles. + + ```python exec="on" + import pandas as pd + import numpy as np + df = pd.read_csv('docs/assets/tables/MIG/llm-finetuning.csv') + df.replace(np.nan, 'NA', inplace=True) + print(df.to_markdown(index=False)) + ``` + + - **Peak Allocated ≈ 5.7 GB across all runs**: The model + LoRA fine-tune has a fixed memory demand, regardless of MIG size. + + - **Peak Reserved varies** (8.9 → 23.5 GB): PyTorch’s caching allocator grabs bigger chunks when more GPU memory is available, but this doesn’t change training feasibility. + + - **Efficiency vs. Speed**: Smaller MIGs (e.g., 10 GB, 20 GB) can be more cost-efficient per token, while larger MIGs or the full 80 GB GPU finish training faster. + + - **Choosing a profile**: The right option depends on priorities — use smaller MIGs to save SUs on long jobs, or larger MIGs when wall-time (speed) is more important. + + !!! info + SU values are calculated as: + `SU = (max(#CPUs, #RAM/4GB) + 16 × (GPU_mem/80)) × hours` + + **Example** (A100_20GB, 0.556 hr walltime, 1 CPU, 4 GB RAM, 20 GB GPU): + ``` + SU = (max(1, 4/4) + 16 × (20/80)) × 0.556 + = (1 + 4) * 0.556 = 2.78 + ``` + +=== "Matrix Multiplication Benchmarks" + + We ran a **matrix multiplication benchmark** on different NVIDIA A100 MIG profiles and the full GPU. The test multiplies large square matrices (sizes like 4096×4096 up to 49k×49k) using PyTorch and CUDA. + + Matrix multiplication is the **core operation in deep learning** — it’s what neural networks spend most of their time doing. Measuring how many **TFLOPs (trillion floating point operations per second)** each MIG slice achieves gives a good picture of its raw compute power. + + ```python exec="on" + import pandas as pd + import numpy as np + df = pd.read_csv('docs/assets/tables/MIG/matrix_multiplication.csv') + df.replace(np.nan, 'NA', inplace=True) + print(df.to_markdown(index=False)) + ``` + + - **Peak FP16 performance** (fast half-precision mode used in AI training). + - **Peak FP32 performance** (single precision with TF32 tensor cores, higher accuracy but slower). + - **Largest tested matrix size (n)** where peak performance was observed. + - **Peak GPU memory usage**, to see whether memory or compute was the bottleneck. + - **SU usage factor**, to tie performance back to billing. + + The results show that **performance scales almost linearly with MIG size (number of SMs)**, while memory never became the limiting factor. This means compute capacity is the main driver of speed, and users can choose between smaller slices (cheaper, slower) or larger slices (faster, higher SU rate) depending on their workload needs. + diff --git a/docs/MIG/profile-comparison.md b/docs/MIG/profile-comparison.md new file mode 100644 index 000000000..e74ad803f --- /dev/null +++ b/docs/MIG/profile-comparison.md @@ -0,0 +1,69 @@ +# MIG Profile Comparison + +MIG profiles represent different partitions of a physical NVIDIA A100 80GB GPU. Each profile gives users a slice of compute and memory resources while maintaining full isolation from other workloads running on the same GPU. + +On Wulver, the following MIG profiles are supported: + +- `10gb` – 10 GB memory +- `20gb` – 20 GB memory +- `40gb` – 40 GB memory + + + +The table below summarizes the hardware characteristics of each MIG profile available on Wulver, alongside the full NVIDIA A100 80 GB GPU. It lists memory capacity, compute resources, and other architectural limits so users can quickly compare performance and capability across profiles. + +```python exec="on" +import pandas as pd +import numpy as np +df = pd.read_csv('docs/assets/tables/MIG/mig-profile-comparison.csv') +df.replace(np.nan, 'NA', inplace=True) +print(df.to_markdown(index=False)) +``` + + \ No newline at end of file diff --git a/docs/OnDemand/2_files.md b/docs/OnDemand/2_files.md new file mode 100644 index 000000000..1847e7cdb --- /dev/null +++ b/docs/OnDemand/2_files.md @@ -0,0 +1,17 @@ +# Files +## Overview + +Files provide you UI based access to your `/home`, `/project` and `/scratch` directory. + +{ width=80% height=80%} + + +## Guide + +{ width=60% height=60%} + +The File Manager tool is available under Files from the Dashboard. Here you can view, edit, and transfer files between your local computer and the cluster. You can access any of the shared filesystems on Wulver including your `$HOME` directory as well as Project, Research, and Scratch. This graphical interface makes it easy to navigate your directories and transfer files to the cluster. (This transfer is only for small files such as job scripts or input scripts, please use command line tools, such as [rsync](https://linux.die.net/man/1/rsync) for larger datasets). + +Use the Upload and Download buttons to transfer files between your local computer and the cluster. You can navigate to any of your directories through the Change Directory button where you can enter the path for your desired location. You can also create new folders with the New Directory button. + + diff --git a/docs/OnDemand/3_clusters.md b/docs/OnDemand/3_clusters.md new file mode 100644 index 000000000..49a51c65b --- /dev/null +++ b/docs/OnDemand/3_clusters.md @@ -0,0 +1,15 @@ +# Clusters +## Overview + +Cluster shell provides you CLI access to Wulver through browser based terminal. + + +{ width=80% height=80%} + + +{ width=80% height=80%} + + + +!!! Tip + If you are on windows and don't have shell access, then this feature is very helpful. diff --git a/docs/OnDemand/4_tools.md b/docs/OnDemand/4_tools.md new file mode 100644 index 000000000..f653ea97c --- /dev/null +++ b/docs/OnDemand/4_tools.md @@ -0,0 +1,77 @@ +# Tools + +## Overview + +The passenger apps (Tools) on OnDemand are some of the easiest and user-friendly ways to monitor key stats related to your account and Wulver in general. + +{ width=60% height=60%} + + +## Joblist + +This is a simple tool to monitor your past jobs as well as the service units you have consumed in a given time period. To use this tool: + +- Click on the Joblist option in the tools drop down menu. + +- Enter the start date and end date between which you want to monitor your past jobs + +{ width=60% height=60%} + +- You will be presented with a table of your jobs with a few job details atop which you find information like service units consumed, your specified date range and the qos. + +{ width=60% height=60%} + + +## Quota Info + +Quota Info is a tool similar to the command “quota_info” on wulver. This tool allows you to see your account information. This information includes: + +- All your accounts +- The total available SUs for each account +- The SUs consumed for each account +- Your storage information for each account for each partition `/project`, `/scratch` +- Your `/home` storage information + +{ width=80% height=80%} + +This is a very good tool for a quick look at your basic account information and is recommended to be used at least once or twice a month to check your SUs as well as `/home` storage. If you find your `/home `is reaching its upper limit you can use another tool called [homespace](#homespace) described below, to have a detailed look at your `/home` usage. + + +## Checkload + +This is a tool which can be used to monitor the CPU load, state, and other parameters on compute nodes across the entire wulver cluster. This tool is not specific to your account. + +- It can be used to check for idle nodes. +- It can also be used to cross check your requested configuration. +- It can also be crudely used to check the load on the cluster + +{ width=80% height=80%} + + +## Homespace + +This is one of the most useful tool to monitor your `/home` partition in detail. This gives you a list of subdirectories and their respective sizes (in MB). You should monitor your `/home` directory size at least twice a month depending on your usage to be sure you are below the upper limit of 50GB. Ideally you should clean your `/home` directory if it reaches 40GB or above. If your `/home` exceeds this limit it severely impacts some operations like creating new conda envs, installing new packages, etc. + +Also an easy way to keep your `/home` below this limit is to move your conda environment and package default directory to your `/project` directory, the details for this can be found [here](../Software/python/conda.md#export-conda-environment) + +!!! Note + + Since it calculates the storage used by each subdirectory and file in your `/home` directory, it may take some time to load the results. + +{ width=80% height=80%} + +## PS + +This is also a very important tool to monitor processes running on your login node usage. + +{ width=80% height=80%} + +!!! Warning + It is strictly advised to not run any resource consuming process on the login node, instead use compute node. The login node is shared between all the wulver users. Using login node for any such process like creating conda env, installing conda packages, running your jobs/programs is strictly not advised. Repeatedly using the login node for such processes will result in strict action. If you find there is high activity by any process on your login node and you are unsure of the cause please email to [hpc@njit.edu](mailto:hpc@njit.edu) with the screenshot of the output of the PS tool. + + +## Qoslist + +QOS is an important flag for any type of job submission. You can check all the available QOS for each of your accounts using this tool. Click [here](../Software/slurm/index.md#partition-use-partition) for details about QOS. + +{ width=80% height=80%} \ No newline at end of file diff --git a/docs/OnDemand/5_My_Interactive_Sessions.md b/docs/OnDemand/5_My_Interactive_Sessions.md new file mode 100644 index 000000000..f2e7cfee9 --- /dev/null +++ b/docs/OnDemand/5_My_Interactive_Sessions.md @@ -0,0 +1,16 @@ +# My Interactive Sessions + +## Overview + +All of your active OnDemand sessions will be shown. + + +{ width=80% height=80%} + + +{ width=80% height=80%} + + +!!! Warning + + Please make sure to delete/close all of your running session after use otherwise it will keep on consuming SU unless low priority is used. \ No newline at end of file diff --git a/docs/OnDemand/6_jobs.md b/docs/OnDemand/6_jobs.md new file mode 100644 index 000000000..65280c20c --- /dev/null +++ b/docs/OnDemand/6_jobs.md @@ -0,0 +1,105 @@ +# Jobs +## Overview + +The Jobs menu on the menu bar includes both the Job Composer and the Active Jobs tools. The Job Composer assists you to set up and submit jobs to the cluster through a graphical interface using file management tools and access to job templates. + +{ width=80% height=80%} + + +## Job Composer + +{ width=60% height=60%} + +### Creating a New Job + +=== "From Default Template" + + - Click New Job > From Default Template + + { width=60% height=60%} + + - A new job (e.g., Simple Sequential Job) will appear in your job list. + + - Click Open Editor to open the directory in the File Editor. + + { width=60% height=60%} + + - Modify the main_job.sh script as needed. + + { width=60% height=60%} + + - Return to the Job Composer and click Job Options to adjust: + - Job Name + - Cluster (ensure it’s set to wulver) + - Account (It will take the default account. If you have or ever had multiple accounts then you have to specify your account) + + { width=60% height=60%} + + - Click Save to apply changes. + + +=== "From Existing Job" + + - Select an existing job from your list. + - Click New Job > From Selected Job. + - This duplicates the job, allowing you to modify it without altering the original. + +=== "From Specified Path" + + - Click New Job > From Specified Path. + - Enter the full path to a directory containing your job script and necessary files. + - This is useful for jobs prepared outside the Job Composer. + +### Editing the Job Script + +1. In the Job Composer, locate your job and click Open Editor under the Submit Script section. +2. Modify the main_job.sh script with your desired commands and SLURM directives. For example: +``` +#SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID +#SBATCH --error=%x.%j.err +#SBATCH --partition=general +#SBATCH --qos=standard +#SBATCH --nodes=1 +#SBATCH --ntasks-per-node=1 +#SBATCH --time=01:00 # D-HH:MM:SS +#SBATCH --mem-per-cpu=4000M +``` +3. Ensure you adjust the SLURM directives (#SBATCH lines) according to your job’s requirements. +4. Click **Save** after editing. + +### Submitting the Job + +1. In the Job Composer, select your job. +2. Click Submit. +3. Monitor the job’s status under the Active Jobs tab. + - Queued: Waiting for resources. + - Running: Currently executing. + - Completed: Finished execution. + +{ width=60% height=60%} + +### Outputs + +- You can check the output/error in folder contents + +{ width=60% height=60%} + +- Check both files to confirm your program ran successfully. + +{ width=60% height=60%} + +!!! Note + Even if an error might have occurred, your job status will still show complete. + + +## Active Jobs + +The Active Jobs tool will allow you to view all the jobs you’ve submitted that are currently in the queue, via OnDemand or not, so you can check on their status. + +{ width=60% height=60%} + +You can expand each job to check more details. You can also open the current working directory of job in file manager or terminal by clicking `Open in File Manger` or `Open in Terminal` respectively. + +{ width=60% height=60%} + + diff --git a/docs/OnDemand/Interactive_Apps/Matlab.md b/docs/OnDemand/Interactive_Apps/Matlab.md new file mode 100644 index 000000000..76bff26a7 --- /dev/null +++ b/docs/OnDemand/Interactive_Apps/Matlab.md @@ -0,0 +1,120 @@ +# MATLAB + +## Overview + +We have three different ways to connect Matlab: + +* Matlab VNC +* Matlab Server +* Jupyter Matlab Proxy + +{ width=30% height=30%} + +## Guide + +=== "Matlab VNC" + + ## Launching MATLAB + + * Navigate to the Interactive Apps section. + * Select `MATLAB` from the list of available applications. + + ## Loading the Matlab Version + + * Select the dropdown option in `MATLAB Version`. The current versions installed on Wulver are `2023a` and `2024a` + + { width=60% height=60%} + + ## Configuring Resources + + * Specify your Slurm account/partition/qos. + * Set the maximum wall time requested. + * Choose the number of cores you need. + * If required, specify the number of GPUs. + + { width=60% height=60%} + + { width=60% height=60%} + + { width=60% height=60%} + + ## Launching the Session + + * Select the `Launch` option after finalizing the resources. Once clicking **Launch**, the request will be queued, and when resources have been allocated, you will be presented with the option to connect to the session by clicking on the blue `Launch MATLAB` option. + + { width=60% height=60%} + + { width=60% height=60%} + + * You might see `Unable to contact settings server` message. It does not mean the Matlab session is terminated. You need to wait a few minutes to see the Matlab popup window. + + + { width=60% height=60%} + + { width=60% height=60%} + + { width=60% height=60%} + +=== "Matlab Server" + + * Select Matlab Server from the interactive apps dropdown menu. + + * Fill in your configurations based on your job requirements + + { width=60% height=60%} + + * Wait for the job to start and then click on **Connect to Matlab** + + { width=60% height=60%} + + * Select existing License + + { width=40% height=40%} + + * Click **Start Matlab** + + { width=40% height=40%} + + * Wait for couple minutes + + { width=40% height=40%} + + * Start working!! + + { width=60% height=60%} + +=== "Jupyter Matlab Proxy" + + + + + * Select **Jupyter-matlab-proxy** from the dropdown menu. + + * Choose matlab-proxy as conda env + + { width=60% height=60%} + + * Fill the rest of the form based on your desired configurations and click Launch. Wait for couple of seconds (same way as other matlab servers) + + * Once the Jupyter opens, click on Open Matlab + + { width=60% height=60%} + + * Select existing License + + { width=40% height=40%} + + * Click start Matlab + + { width=40% height=40%} + + * Wait for couple minutes + + { width=40% height=40%} + + * Start working!! + + { width=60% height=60%} + + + diff --git a/docs/OnDemand/Interactive_Apps/Notebook.md b/docs/OnDemand/Interactive_Apps/Notebook.md new file mode 100644 index 000000000..98f84a14c --- /dev/null +++ b/docs/OnDemand/Interactive_Apps/Notebook.md @@ -0,0 +1,42 @@ +# Jupyter + +## Launching Jupyter Session + +* Navigate to the Interactive Apps section. +* Select `Jupyter` from the list of available applications. + +## Loading the Environment + +* Choose the `Mode` option, where you can select the interface: + - Jupyterlab + - Jupyter Notebook + +* In the `Conda Environment` section, you can see all your conda environments automatically detected, please select the one which you want to work with. + +!!! Note + Please make sure that Jupyter package is installed in your conda env otherwise you will get an error. If you are unsure how to install Jupyter Notebook or Jupyterlab in the Conda environment, check [Conda Documentation](conda.md) and [Jupyter Installation](jupyter.md). + +* Choose the path where you want to start the Jupyter session in `Enter the full path of the case directory`. For session in `$HOME` directory keep this blank. + +{ width=60% height=60%} + +## Configuring Resources + +* Specify your Slurm account/partition/qos. +* Set the maximum wall time requested. +* Choose the number of cores you need. +* If required, specify the number of GPUs. + +{ width=60% height=60%} + +{ width=60% height=60%} + + +## Launching the Session + +Select the `Launch` option after finalizing the resources. Once clicking **Launch**, the request will be queued, and when resources have been allocated, you will be presented with the option to connect to the session by clicking on the blue `Connect to Jupyter` option. + +{ width=60% height=60%} + +{ width=60% height=60%} + diff --git a/docs/OnDemand/Interactive_Apps/Rstudio.md b/docs/OnDemand/Interactive_Apps/Rstudio.md new file mode 100644 index 000000000..bdfad6336 --- /dev/null +++ b/docs/OnDemand/Interactive_Apps/Rstudio.md @@ -0,0 +1,49 @@ +# RStudio + + + +## Launching RStudio + +* Navigate to the Interactive Apps section. +* Select `RStudio` from the list of available applications. + +## Loading the R Environment + +* Please select your desired R version from the drop down. +* To use a different version or environment, please select "custom" and enter the necessary commands: +For example, to use a Conda environment, enter: +`module load Anaconda3/2023.09-0; source conda.sh; conda activate my_conda_r_env` + + +{ width=50% height=50%} + +## Configuring Resources + +* Specify your Slurm account/partition/qos. +* Set the maximum wall time requested. +* Choose the number of cores you need. +* If required, specify the number of GPUs. + +## Launching the Session + +{ width=40% height=20%} + +Once clicking **Launch**, the request will be queued, and when resources have been allocated, you will be presented with the option to connect to the session by clicking on the blue Connect to R Studio Server button. +{ width=50% height=50%} + +{ width=50% height=50%} + +{ width=50% height=50%} + +Once connected, the familiar R Studio interface is presented, and you will be able to use the allocated resources, and access your research data located on Wulver. +Installing packages +It's likely your scripts will require additional R libraries; these can be installed using the `install.packages()` command in the console window of R Studio. + +## Exiting the session +If a session exceeds the requested running time, it will be killed. You may receive a message "The previous R session was abnormally terminated...". Click OK to acknowledge the message and continue. To avoid this message, it's good practice to exit the session cleanly when you have finished. +To cleanly exit R Studio, click `File -> Quit Session...` and then release resources back to the cluster queues by clicking the red Delete button for the relevant session on the My Interactive Sessions page. + + + + + diff --git a/docs/OnDemand/Interactive_Apps/index.md b/docs/OnDemand/Interactive_Apps/index.md new file mode 100644 index 000000000..3171b3ea9 --- /dev/null +++ b/docs/OnDemand/Interactive_Apps/index.md @@ -0,0 +1,13 @@ +# Interactive Apps + +## Overview + +We have a lot of interactive apps which you can use for UI interface. They run as jobs in similar way as you would do using shell, except these are interactive. For example, you can start Jupyter Notebook app and work on it. Your files will be saved in directory created by the ondemand unless you specify a different directory during the configurations. + + +{ width=80% height=80%} + + +!!! Warning + Please make sure to close your app session once you have completed your work otherwise it will keep consuming SUs. + diff --git a/docs/OnDemand/index.md b/docs/OnDemand/index.md new file mode 100644 index 000000000..ac2afebd9 --- /dev/null +++ b/docs/OnDemand/index.md @@ -0,0 +1,34 @@ +# Open OnDemand + +## Overview + +Open OnDemand is now available for NJIT HPC access at [ondemand.njit.edu](https://ondemand.njit.edu)! + +[Open OnDemand](https://openondemand.org/) is a browser-based gateway to NJIT's Wulver cluster and shared storage. It offers a graphical interface allowing users to view, edit, download, and upload files. Users can manage and create job templates for the cluster and access interactive applications such as remote desktops to cluster nodes. Additionally, Open OnDemand supports GUI-based software like Jupyter Lab/Notebook, Matlab, and RStudio, all accessible through a web browser on virtually any device. No additional software installation is necessary, and users can operate with minimal Linux and job scheduler command knowledge. + + +This is an [open source](https://openondemand.org/) project developed through NSF funding. + +## Features + +* Easy to use +* Great for researchers and students new to HPC +* Convenient for experienced users as well + +## Using OnDemand + + +### Logging into OnDemand + +{ width=60% height=60%} + +If you have access to the Wulver cluster, you can use OnDemand. Open any browser and go to ondemand.njit.edu. Use your UCID and password to log in. If you are off campus, you will need to set up VPN to access the platform. + + +### Dashboard + +{ width=60% height=60%} + +Once you log in, you will see the OnDemand Dashboard. You will see the menu bar on the top where you can access all the tools available including Files Manager, Shell Access, Job Composer, and Interactive Apps. You will also see several pinned apps highlighted on the Dashboard. + + diff --git a/docs/Policies/condo_policies.md b/docs/Policies/condo_policies.md new file mode 100644 index 000000000..ea7c0fa6e --- /dev/null +++ b/docs/Policies/condo_policies.md @@ -0,0 +1,37 @@ +# Shared Condo Partnership + +## Cluster Resource Investment and Priority Access Policy + +Faculty members who regularly require more resources than the standard allocation may choose to invest in additional resources—either partial or full nodes—thereby contributing to the growth of the cluster. A catalog of available partial node and GPU investment options is provided below. Principal Investigators (PIs) and their associated users will receive higher job priority, up to the amount of resources they have contributed. Please note that jobs may not necessarily run on the specific nodes purchased; instead, the contributed resources will be made available through a floating reservation equivalent to the purchased capacity. + +Contributors can also submit jobs using standard SUs and at lower priority (See [Job QoS](node-memory-config.md)) beyond their reserved allocation. The university will cover all infrastructure-related costs for these contributed nodes. This floating reservation will remain in effect for **five years**. If the hardware is upgraded or replaced before the end of this period, the job priority will transfer to the closest equivalent resources on the new hardware. + +## Full node investment +Please contact [hpc@njit.edu](mailto:hpc@njit.edu) to discuss your specific computational needs. + +## Partial Node Investment +You can invest in partial nodes, either on a per-CPU or per-MIG GPU basis. This flexible model allows you to customize and build resources tailored to your research requirements. Rates for HPC resources are provided below to help you plan your investment. + +```python exec="on" +import pandas as pd +df = pd.read_csv('docs/assets/tables/condo.csv') +print(df.to_markdown(index=False)) +``` + +!!! info + + MIG (Multi-Instance GPU) allows a single NVIDIA GPU (like the A100) to be split into multiple independent instances, each with dedicated compute and memory resources. This enables multiple users to share a GPU efficiently. It’s ideal for running smaller workloads without needing a full GPU. + +**Example:** If your research workflow requires 128 CPU cores with 4 GB RAM per core, one 40 GB MIG, and one 20 GB MIG, you can invest in 128 CPUs at $150 per core ($19,200), plus the MIGs ($5,000 for 40 GB and $2,500 for 20 GB), for a total cost of $26,700. + +!!! tips + + Please contact [hpc@njit.edu](mailto:hpc@njit.edu) with an index to invest in partial nodes. Note that all invoices get billed to HPC Services account and not the Equipment account. + +## Dedicated Pool + +If the shared condo module does not satisfy the needs of the PI, a dedicated pool may be set up. In addition to the nodes, the PI will be charged for all infrastructure costs, including but not limited to electricity, HVAC, system administration, etc. It is strongly recommended to first try the shared condo model. If the shared condo model does not work, the nodes can be converted to a dedicated pool. + +!!! warning + + A dedicated pool of partial node or gpus is not available, only full node investments are allowed. diff --git a/docs/Policies/index.md b/docs/Policies/index.md new file mode 100644 index 000000000..07796ba89 --- /dev/null +++ b/docs/Policies/index.md @@ -0,0 +1,7 @@ +# Wulver Usage and Condo Policies +All users must be associated with a Principal Investigator's (PI's) allocation. Course-based allocations expire at the end of the semester. If you have accessed Wulver as part of a course, you will need to be associated with a PI's research allocation to retain access after the semester concludes. + +See the following details on Wulver Usage and Condo Policies + +* [Wulver Polices](wulver_policies.md) +* [Condo Polices](condo_policies.md) \ No newline at end of file diff --git a/docs/Policies/wulver_policies.md b/docs/Policies/wulver_policies.md index 01d3c1e51..19b455414 100644 --- a/docs/Policies/wulver_policies.md +++ b/docs/Policies/wulver_policies.md @@ -1,39 +1,31 @@ -# Wulver Usage and Condo Policy - -Proposed Wulver Usage and Condo Policies - -These policies are considered draft. We will work with faculty and senior administration to fine tune these policies. There are still many details to be worked out. We welcome comments. +# Wulver Policies ## Faculty Computing Allowance -Faculty PIs are allocated 300,000 Service Units (SU) per year on request at no cost. An SU is equal to 1 core hour on a standard node. More details on calculating SUs for GPUs, high memory nodes, etc… will be provided at a later date. All users working as part of the PIs project will use this allocation. Multiple PIs working on the same project may pool SUs. The SUs can be renewed annually by providing a brief report describing how the SUs were used and list of publications, presentations, and awards generated from research conducted. Additional SUs may be purchased at a cost of $0.01/SU. The minimum purchase is 50,000 SU ($500). - -## User Storage Allowance - -Users will be provided with 50GB home directories. Home directories are backed up. PIs are additionally provided 2TB project directories. These project directories are not backed up. Very fast NVME scratch is available to users. This scratch space is for temporary files generated during a run and will be deleted after the job is complete. Additional project storage can be purchased if needed. This additional project space is not backed up, however, if backup is desired arrangements can be made at additional cost. Costs will be provided. It is important for users to understand the Wulver is a compute device and not a long term storage device. Users need to manage data so that backed up data fits in home directory space. Transient, or rapidly changing data should be stored in the project directory. Long term storage with backups or archival storage for research data will be stored in a yet to be determined campus wide storage resource. +Faculty PIs are allocated 300,000 Service Units (SU) per year on request at no cost. An SU is equal to 1 core hour on a standard node. For more details on calculating SUs for GPUs, see [Service Units](service-units.md). All users working as part of the PIs project will use this allocation. Multiple PIs working on the same project may pool SUs. The SUs can be renewed annually by providing a brief report describing how the SUs were used and a list of publications, presentations, and awards generated from research conducted. Additional SUs may be purchased at a cost of $0.005/SU. The minimum purchase is 50,000 SU (250 USD). +!!! note -## Shared Condo Partnership + The 300,000 SUs are available on `--qos=standard` only. If PI does not want to buy more SUs, PI's group members can use `--qos=low` which does not have any SU charges. For more details, see [SLURM QOS](node-memory-config.md). -Faculty who routinely need more resources than the initial allocation may buy nodes and contribute to the cluster. A catalog of select hardware for inclusion in the cluster will be made available. The PI and associated users will be able to submit jobs with a higher priority up to the resources contributed. Note that these jobs may or may not run on the actual nodes purchased. The allocated resources will be available via a floating reservation for the amount of resources purchased. Contributors will be able to additionally submit jobs using SUs as well as lower priority. The university will subsidize all infrastructure costs for these nodes. This floating reservation will be available for five years. - -## Private Pool +## User Storage Allowance -If the shared condo module does not satisfy the needs of the PI, a private pool may be set up. In addition to the nodes, the PI will be charged for all infrastructure costs, including but not limited to electricity, HVAC, system administration, etc…. It is strongly recommended to first try the shared condo model. If the shared condo model does not work, the nodes can be converted to a private pool. +Users will be provided with 50GB of `$HOME` directories. Home directories are backed up. PIs are additionally provided 2TB project directories. These project directories are backed up. Very fast NVME scratch is available to users. This scratch space is for temporary files generated during a run and will be deleted after 30 days. Additional project storage can be purchased if needed. This additional project space will also be backed up. Users need to manage data so that backed-up data fits in the project directory space. Transient, or rapidly changing data should be stored in the scratch directory. Long-term storage with backups or archival storage for research data will be stored in a yet to be determined campus wide storage resource. See [Wulver Filesystems](Wulver_filesystems.md) for details. ## Job Priorities * Standard Priority - * Charged against the SU allocation. - * Wall time maximum - 72 hours. - * Jobs are not preemptable. + * Faculty PIs are allocated 300,000 Service Units (SU) per year on request at no cost + * Wall time maximum - 72 hours + * Additional SUs may be purchased at a cost of $0.005/SU + * The minimum purchase is 50,000 SU ($250) + * Jobs can be superseded by those with higher priority jobs + * Low Priority * Not charged against SU allocation * Wall time maximum - 72 hours - -Jobs are preemptable by higher priority jobs + * Jobs can be preempted by those with higher and standard priority jobs when they are in the queue * High Priority * Not charged against SU allocation - * Wall time maximum is determined by PI; 3 days are suggested. ARCS reserves right to reboot nodes once a month for maintenance (perhaps first Monday of the month) - * Jobs are not preemptable + * Wall time: 72 hours (default), PI can request longer walltimes up to 30 days. ARCS HPC reserves the right to reboot nodes once a month for maintenance — the second Tuesday of each month. See [Cluster Maintenance Updates and News](../news/index.md) for details * Only available to contributors \ No newline at end of file diff --git a/docs/Running_jobs/array-jobs.md b/docs/Running_jobs/array-jobs.md new file mode 100644 index 000000000..a8dc52728 --- /dev/null +++ b/docs/Running_jobs/array-jobs.md @@ -0,0 +1,90 @@ +## Overview + +Array jobs allow you to submit many similar jobs (like simulations or data processing tasks) with a single sbatch command. Each job in the array runs the same script but can process different input parameters. + +**Analogy:** Imagine you're baking cookies in multiple batches. Each tray (array job) uses the same recipe (your script), but maybe with a different flavor (input). Instead of submitting each tray separately, you give the oven a list and it bakes them one by one or in parallel! + +### Why use Array jobs? +Array jobs are powerful and efficient for batch-processing many similar tasks. They save time, simplify management, and optimize cluster usage. + +If you’re running `50` experiments with the same script — don’t submit `50` jobs. Use an array job instead! + +- Simplifies the submission of multiple similar jobs with a single script +- Reduces scheduler overhead by bundling related tasks into one job +- Keeps your job queue cleaner and more organized +- Makes it easier to monitor, debug, and manage large-scale workflows +- Ideal for training machine learning models on multiple datasets +- Useful for running simulations across a range of input parameters +- Efficient for processing large datasets by splitting them into manageable chunks + +## Special Variables in Array Jobs + +| Variable | Description | +|---------------------------|--------------------------------------------------| +| `SLURM_JOB_ID` | Unique ID for each job element (individual task) | +| `SLURM_ARRAY_JOB_ID` | Shared job ID for the entire job array | +| `SLURM_ARRAY_TASK_ID` | The task ID for the current job in the array | +| `SLURM_ARRAY_TASK_MIN` | The lowest task ID in this job array | +| `SLURM_ARRAY_TASK_MAX` | The highest task ID in this job array | +| `SLURM_ARRAY_TASK_COUNT` | Total number of tasks in the job array | + + +## Array Job Examples +```slurm +#!/bin/bash -l +#SBATCH -J myprogram +#SBATCH --partition=general +#SBATCH --qos=standard +#SBATCH --nodes=1 +#SBATCH --ntasks-per-node=1 +#SBATCH --array=1-30 +#SBATCH --output=myprogram%A_%a.out +#SBATCH --error=myprogram%A_%a.err +#SBATCH --time=71:59:59 + +./myprogram input$SLURM_ARRAY_TASK_ID.dat +sleep 10 +``` + +This example demonstrates how to use SLURM job arrays to run the same program multiple times with varying inputs. Here's what each part of the script does: + +- `#SBATCH --array=1-30`: This line creates a job array with 30 tasks. Each task in the array gets a unique `SLURM_ARRAY_TASK_ID` from 1 to 30. + +- `#SBATCH --output=myprogram%A_%a.out` +`#SBATCH --error=myprogram%A_%a.err`: These lines set up output and error file names for each array task. %A is replaced with the array job ID. %a is replaced with the task index (from 1 to 30). This prevents files from different tasks from overwriting each other. + +- `./myprogram input$SLURM_ARRAY_TASK_ID.dat`: This runs the program using different input files for each task. For example: + - Task 1 runs: `./myprogram input1.dat` + - Task 2 runs: `./myprogram input2.dat` + - …and so on up to `input30.dat` + + +- `sleep 10`: This is just a placeholder command to simulate a small wait time. You can remove or replace it as needed. + + +## Job Array Use Case + +I have an application, app, that needs to be run against every line of my dataset. Every line changes how app runs slightly, but I need to compare the runs against each other. + +Older, slower way of homogenous batch submission: + +```shell +#!/bin/bash +DATASET=dataset.txt +scriptnum = 0 +while read LINE; do +echo "app $LINE" > ${scriptnum}.sh +sbatch ${scriptnum}.sh +scriptnum=$(( scriptnum + 1 )) +done < $DATASET +``` + +Not only is this needlessly complex, it is also slow, as sbatch has to verify each job as it is submitted. This can be done easily with array jobs, as long as you know the number of lines in the dataset. This number can be obtained like so: `wc -l` dataset.txt in this case lets call it `100`. + +Better way: + +```slurm +#!/bin/bash +#SBATCH - - array=1-100 +srun app `sed - n "${SLURM_ARRAY_TASK_ID}"` dataset.txt +``` \ No newline at end of file diff --git a/docs/Running_jobs/batch-jobs.md b/docs/Running_jobs/batch-jobs.md new file mode 100644 index 000000000..1127e396a --- /dev/null +++ b/docs/Running_jobs/batch-jobs.md @@ -0,0 +1,223 @@ +## Overview +Batch jobs are like pre-written instructions handed over to the cluster to be executed when resources become available. + +Unlike your personal laptop where you run commands interactively, HPC jobs are queued and run asynchronously using a job script — a text file that tells Slurm: + +- What resources do you need? +- What is your PI account? +- What programs or commands are needed to run job? +- How long your job may take? + +### Example of batch job slurm script + +=== "CPU Nodes" + ??? example "Sample Job Script to use: submit.sh" + + === "Using 1 core" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=job_name + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks=1 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + ./myexe # myexe is the executable in this example. + ``` + + === "Using multiple cores" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=job_name + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + srun ./myexe # myexe is the executable in this example. + ``` + + === "Using multiple threads" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=job_name + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + OMP_NUM_THREADS=$SLURM_NTASKS ./myexe + ``` + Use this script, if your code relies on threads instead of cores. + + === "Using multiple cores and threads" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=job_name + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks=64 + #SBATCH --cpus-per-task=2 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + srun gmx_mpi mdrun ... -ntomp $SLURM_CPUS_PER_TASK ... + ``` + This is the example script of [GROAMCS](gromacs.md) which uses both CPUs and threads. + !!! warning + + Do not request multiple cores unless your code is parallelized. Before using multiple cores, ensure that your code is capable of parallelizing tasks; otherwise, it will unnecessarily consume service units (SUs) and may negatively impact performance. Please review the code's documentation thoroughly and use a single core if it does not support parallel execution. + + * Here, the job requests 1 node on the `general` partition with `qos=standard`. Please note that the memory relies on the number of cores you are requesting. + * As per the policy, users can request up to 4GB memory per core, therefore the flag `--mem-per-cpu` is used for memory requirement. If you are using 1 core and need more memory, use `--mem` instead. + * In this above script `--time` indicates the wall time which is used to specify the maximum amount of time that a job is allowed to run. The maximum allowable wall time depends on SLURM QoS, which you can find in [QoS](node-memory-config.md)). + * To submit the job, use `sbatch submit.sh` where the `submit.sh` is the job script. Once the job has been submitted, the jobs will be in the queue, which will be executed based on priority-based scheduling. + * To check the status of the job use `squeue -u $LOGNAME` and you should see the following + ```bash + JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON) + 635 general job_nme ucid R 00:02:19 1 n0088 + ``` + Here, the `ST` stands for the status of the job. You may see the status of the job `ST` as `PD` which means the job is pending and has not been assigned yet. The status change depends upon the number of users using the partition and resources requested in the job. Once the job starts, you will see the output file with an extension of `.out`. If the job causes any errors, you can check the details of the error in the file with the `.err` extension. + +=== "GPU Nodes" + In case of submitting the jobs on GPU, you can use the following SLURM script + + ??? example "Sample Job Script to use: gpu_submit.sh" + + === "Using 1 core, 1 GPU" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=gpu_job + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=1 + #SBATCH --gres=gpu:1 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + ./myexe # myexe is the executable in this example. + ``` + + === "Using multiple cores, 1 GPU" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=gpu_job + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --gres=gpu:1 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + srun ./myexe # myexe is the executable in this example. + ``` + + === "Using multiple cores, GPUs" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=gpu_job + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --gres=gpu:2 + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + srun ./myexe # myexe is the executable in this example. + ``` + + === "Using MIGs" + ```slurm + #!/bin/bash -l + #SBATCH --job-name=gpu_job + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --gres=gpu:a100_10g:1 # This uses 10G MIG, to use 20G or 40G MIG, modify 10g to 20g or 40g + #SBATCH --time=59:00 # D-HH:MM:SS + #SBATCH --mem-per-cpu=4000M + + srun ./myexe # myexe is the executable in this example. + ``` + + !!! warning + + Do not use multiple GPUs unless you are certain that your job's performance will benefit from them. Most GPU jobs do not require multiple CPUs either. Please remember that unnecessarily requesting additional resources can negatively impact job performance and will also consume more service units (SUs). + +=== "Debug Node" + The `debug` QoS in Slurm is intended for debugging and testing jobs. It usually provides a shorter queue wait time and quicker job turnaround. Jobs submitted with the `debug` QoS have access to a limited set of resources (Only 4 CPUS on Wulver), making it suitable for rapid testing and debugging of applications without tying up cluster resources for extended periods. + + ??? example "Sample Job Script to use: debug_submit.sh" + + ```slurm + #!/bin/bash -l + #SBATCH --job-name=debug + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=debug + #SBATCH --qos=debug + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=1 + #SBATCH --time=7:59:00 # D-HH:MM:SS, Maximum allowable Wall Time 8 hours + #SBATCH --mem-per-cpu=4000M + + ./myexe + ``` +=== "Bigmem Node" + The bigmem nodes provide 2 TB of RAM in total. By default, each CPU core is allocated 16 GB of memory, but you can request additional memory if your job requires it. + + ??? example "Sample Job Script to use: bigmem_submit.sh" + + ```slurm + #!/bin/bash -l + #SBATCH --job-name=bigmem_job + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --partition=bigmem + #SBATCH --qos=bigmem + #SBATCH --account=$PI_ucid # Replace $PI_ucid which the NJIT UCID of PI + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=1 + #SBATCH --time=59:00 # D-HH:MM:SS, Maximum allowable Wall Time 8 hours + #SBATCH --mem-per-cpu=16000M + + ./myexe + ``` + +To submit the jobs, `sbatch` command. \ No newline at end of file diff --git a/docs/Running_jobs/checkpointing.md b/docs/Running_jobs/checkpointing.md new file mode 100644 index 000000000..f9eedb422 --- /dev/null +++ b/docs/Running_jobs/checkpointing.md @@ -0,0 +1,139 @@ +## Overview + +Checkpointing is the process of saving the current state of a running job at regular intervals so that it can be resumed later from that state, rather than starting from scratch. This is especially useful in long-running or resource-intensive tasks on HPC systems like Wulver, where interruptions or failures may occur. + +Checkpointing typically involves: + +- Periodic saving of application state (memory, variables, file handles, etc.) +- Resuming computation from the last saved state +- Integration with SLURM job re-submission or recovery workflows + +## Why Use Checkpointing? + +| Benefit | Description | +|--------|-------------| +| **Failure Recovery** | Resume jobs from the last checkpoint after a node crash or time expiration. | +| **Efficient Resource Use** | Prevents waste of computation time on long jobs that are interrupted. | +| **Preemption Tolerance** | Helps tolerate job preemption on shared clusters or spot instances. | +| **Job Time Limit Bypass** | Breaks large jobs into smaller chunks to fit within SLURM time limits. | + + +## Examples for checkpointing + +=== "Python" + + Save intermediate state using Python’s built-in `pickle` module — ideal for lightweight scripts. + + ```python + import pickle + import time + + def save_checkpoint(data, filename="checkpoint.pkl"): + with open(filename, "wb") as f: + pickle.dump(data, f) + + def load_checkpoint(filename="checkpoint.pkl"): + try: + with open(filename, "rb") as f: + return pickle.load(f) + except FileNotFoundError: + return {"iteration": 0} + + state = load_checkpoint() + for i in range(state["iteration"], 1000): + # Do some work + time.sleep(1) + print(f"Running step {i}") + + # Save progress every 100 steps + if i % 100 == 0: + save_checkpoint({"iteration": i}) + + ``` + +=== "Pytorch" + + A common practice in PyTorch to checkpoint model weights, optimizer state, and epoch index — useful for training recovery. + + ```python + import torch + + # Save checkpoint + torch.save({ + 'epoch': epoch, + 'model_state_dict': model.state_dict(), + 'optimizer_state_dict': optimizer.state_dict() + }, 'checkpoint.pth') + + # Load checkpoint + checkpoint = torch.load('checkpoint.pth') + model.load_state_dict(checkpoint['model_state_dict']) + optimizer.load_state_dict(checkpoint['optimizer_state_dict']) + epoch = checkpoint['epoch'] + ``` + +=== "Tensorflow" + + Using Keras callbacks, checkpoints are saved automatically during training. Only model weights are saved to keep storage efficient. + + ```python + import tensorflow as tf + + model = tf.keras.models.Sequential([...]) + checkpoint_path = "checkpoints/model.ckpt" + checkpoint_cb = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path, + save_weights_only=True, + save_freq='epoch') + + model.fit(data, labels, epochs=10, callbacks=[checkpoint_cb]) + ``` + +=== "C/C++" + + In C/C++, you can implement basic checkpointing by writing a loop index or state to a file and loading it at the next run. Ideal for simple simulations or compute-intensive loops. + + ```cpp + int main() { + int current_step = load_checkpoint("state.dat"); // Custom function + for (int i = current_step; i < MAX; i++) { + // Work + if (i % 1000 == 0) { + save_checkpoint(i, "state.dat"); + } + } + } + ``` + +=== "GROMACS" + + GROMACS supports checkpointing with `.cpt` files during molecular simulations: + + ```shell + gmx mdrun -deffnm simulation -cpt 15 # Saves checkpoints every 15 minutes + + # Restart from checkpoint + gmx mdrun -s topol.tpr -cpi simulation.cpt + ``` + +=== "LAMMPS" + + LAMMPS checkpointing is usually done using `write_restart` and `read_restart`: + + ```lammps + write_restart restart.equilibration + + # In a new job + read_restart restart.equilibration + ``` + +=== "OpenFOAM (CFD)" + + Checkpointing is done by writing time steps to disk and restarting from a previous time directory: + + ```shell + # Set in controlDict + writeInterval 100; + + # Restart + startFrom latestTime; + ``` diff --git a/docs/Running_jobs/dependency-jobs.md b/docs/Running_jobs/dependency-jobs.md new file mode 100644 index 000000000..5f1cb7040 --- /dev/null +++ b/docs/Running_jobs/dependency-jobs.md @@ -0,0 +1,57 @@ +## Overview + +In many workflows, one job must start only after another has successfully completed. For example, you might want to: + +- Preprocess data in one job and then analyze it in another +- Run a simulation, then run a visualization job +- Compile a program, and then run the executable + +Slurm allows you to chain jobs together using job dependencies, so that one job begins only when the specified condition on another job is met. + +This avoids manual tracking and reduces errors in job sequencing. + +## Job Dependency Options + +| Dependency Type | Description | +|---------------------------------------|-------------| +| `after:job_id[:job_id...]` | This job can begin execution **after the specified jobs have started**. | +| `afterany:job_id[:job_id...]` | This job can begin execution **after the specified jobs have terminated**, regardless of state. | +| `afterburstbuffer:job_id[:job_id...]` | This job starts **after the specified jobs have terminated** and any associated **burst buffer stage-out operations** are complete. | +| `afternotok:job_id[:job_id...]` | This job starts **only if the specified jobs fail** (non-zero exit code, node failure, timeout, etc). | +| `afterok:job_id[:job_id...]` | This job starts **only if the specified jobs succeed** (exit code 0). | +| `aftercorr:job_id` | Each task in this job array will start **after the corresponding task ID** in the specified job has completed successfully. | +| `singleton` | This job will only start **after any previous jobs with the same name and user have finished**. | + +## Job Dependency Examples + +**Run Second Job After First Job Completes Successfully** + +```shell +# Submit first job +$ sbatch preprocess.sh +Submitted batch job 12345 +``` + +```shell +# Submit dependent job (afterok = only if first job succeeds) +$ sbatch --dependency=afterok:12345 analyze.sh +Submitted batch job 12346 +``` + +In this example: + +- `12345` is the job ID of the first job. +- The second job will start only if preprocess.sh exits with code 0 (success). + +**Chaining Multiple Jobs** + +```shell +# Submit step 1 +$ sbatch step1.sh # returns JobID 11111 + +# Submit step 2 to run after step 1 +$ sbatch --dependency=afterok:11111 step2.sh # returns JobID 11112 + +# Submit step 3 to run after step 2 +$ sbatch --dependency=afterok:11112 step3.sh +``` diff --git a/docs/Running_jobs/index.md b/docs/Running_jobs/index.md new file mode 100644 index 000000000..f189a0d2c --- /dev/null +++ b/docs/Running_jobs/index.md @@ -0,0 +1,42 @@ +# Overview + +Wulver is a shared resource among researchers, faculty and students. It is important to use it efficiently so that everyone can complete their tasks without delay. Therefore, running jobs on Wulver, you should follow certain norms which ensures that your work is done on time and also lets others run their task without any conflict. We use Slurm on Wulver to schedule and manage jobs. + + +## What is SLURM? + +Slurm (Simple Linux Utility for Resource Management) is an open-source workload manager and job scheduler designed for high-performance computing clusters. It is widely used in research, academia, and industry to efficiently manage and allocate computing resources such as CPUs, GPUs, memory, and storage for running various types of jobs and tasks. Slurm helps optimize resource utilization, minimizes job conflicts, and provides a flexible framework for distributing workloads across a cluster of machines. It offers features like job prioritization, fair sharing of resources, job dependencies, and real-time monitoring, making it an essential tool for orchestrating complex computational workflows in diverse fields. + + +## Some best practices to follow: + +- **Request Only the Resources You Need :** + Be precise when requesting CPUs, memory, GPUs, and runtime.
+ Avoid overestimating job time (`--time`) and memory (`--mem`) as it reduces scheduler efficiency.
+ Use monitoring tools to understand your typical usage patterns and adjust accordingly.
+ +- **Do not run jobs on Login Node :** + Login node is the entry point for Wulver and has limited memory and resources.
+ Please avoid directly running jobs on the login node as it can slow down the system for everyone.
+ Always submit jobs to compute nodes via slurm script or start an interactive session. + +- **Use Appropriate Partitions :** + Submit jobs to the correct partition based on resource needs (e.g., GPU, high-memory).
+ +- **Test and Debug with Small Jobs First :** + Use short test runs or dedicated debug partitions for code testing or troubleshooting.
+ This helps prevent long-running failures and wasted compute hours.
+ +- **Monitor Your Jobs :** + Please use commands like `squeue`, `slurm_jobid $jobid`, `seff $jobid` to check your job status
+ You can also use our [Ondemand Tools](../OnDemand/4_tools.md).
+ +- **Respect Fair Usage Policies :** + Do not monopolize shared resources by submitting excessive large jobs.
+ Be mindful of Wulver's [usage policy](../Policies/wulver_policies.md).
+ +- **Leverage MIGs for Efficient GPU Utilization :** + Our Nvidia A100 GPUs have MIG implementation which allows a single GPU to be split into multiple isolated instances.
+ Use MIG-compatible partitions when your task doesn’t require the full GPU power
+ More info about [MIG](../MIG/index.md).
+ diff --git a/docs/Running_jobs/interactive-jobs.md b/docs/Running_jobs/interactive-jobs.md new file mode 100644 index 000000000..b3ba236ab --- /dev/null +++ b/docs/Running_jobs/interactive-jobs.md @@ -0,0 +1,92 @@ +## Overview +Interactive jobs allow users to directly access a compute node in real time — as if you were working on your personal computer, but with the power of HPC behind it. + +Unlike batch jobs, which are queued and run in the background, interactive jobs open a live terminal session on a compute node. This is useful for: + +- Testing code or software modules +- Debugging runtime issues +- Running Jupyter notebooks +- Exploring the system environment or dependencies + +You still need to request resources via Slurm, but instead of submitting a script with sbatch, you request an interactive shell using our `interative` command. + +#### The `interactive` Command +We provide a built-in shortcut command, `interactive`, that allows you to quickly and easily request a session in compute node. + +The `interactive` command acts as a convenient wrapper for Slurm’s [salloc](https://slurm.schedmd.com/salloc.html) command. Similar to [sbatch](https://slurm.schedmd.com/sbatch.html), which is used for batch jobs, `salloc` is specifically designed for interactive jobs. + +```bash +$ interactive -h +Usage: interactive -a ACCOUNT -q QOS -j JOB_TYPE +Starts an interactive SLURM job with the required account and QoS settings. + +Required options: + -a ACCOUNT Specify the account to use. + -q QOS Specify the quality of service (QoS). + -j JOB_TYPE Specify the type of job: 'cpu' for CPU jobs or 'gpu' for GPU jobs. + +Example: Run an interactive GPU job with the 'test' account and 'test' QoS: + /apps/site/bin/interactive -a test -q test -j gpu + +This will launch an interactive job on the 'gpu' partition with the 'test' account and QoS 'test', +using 1 GPU, 1 CPU, and a walltime of 1 hour by default. + +Optional parameters to modify resources: + -n NTASKS Specify the number of CPU tasks (Default: 1). + -t WALLTIME Specify the walltime in hours (Default: 1). + -g GPU Specify the number of GPUs (Only for GPU jobs, Default: 1). + -p PARTITION Specify the SLURM partition (Default: 'general' for CPU jobs, 'gpu' for GPU jobs). + +Use '-h' to display this help message. +``` + +=== "CPU Nodes" + + ```bash + $ interactive -a $PI_UCID -q standard -j cpu + Job Type: cpu + Starting an interactive session with the general partition and 1 core for 01:00:00 of walltime in standard priority + srun: job 584280 queued and waiting for resources + srun: job 584280 has been allocated resources + ``` + +=== "GPU Nodes" + + ```bash + $ interactive -a $PI_UCID -q standard -j gpu + Job Type: gpu + Starting an interactive session with the GPU partition, 1 core and 1 GPU for 01:00:00 of walltime in standard priority + srun: job 584279 queued and waiting for resources + srun: job 584279 has been allocated resources + ``` + +=== "Debug Nodes" + + ```bash + $ interactive -a $PI_UCID -q debug -j cpu -p debug + Job Type: cpu + Starting an interactive session with the debug partition and 1 core for 01:00:00 of walltime in debug priority + srun: job 584281 queued and waiting for resources + srun: job 584281 has been allocated resources + ``` + +Replace `$PI_UCID` with PI's NJIT UCID. +Now, once you get the confirmation of job allocation, you will be assigned to a compute node. + +#### Customizing Your Resources +Please note that, by default, this interactive session will request 1 core (for CPU jobs), 1 GPU (for GPU jobs), with a 1-hour walltime. To customize the resources, use the `-h` option for help. Run `interactive -h` for more details. Here is an explanation of each flag given below. + +```python exec="on" +import pandas as pd +import numpy as np +df = pd.read_csv('docs/assets/tables/interactive.csv') +df.replace(np.nan, 'NA', inplace=True) +print(df.to_markdown(index=False)) +``` +!!! warning + + Login nodes are not designed for running computationally intensive jobs. You can use the head node to edit and manage your files, or to run small-scale interactive jobs. The CPU usage is limited per user on the head node. Therefore, for serious computing either submit the job using `sbatch` command or start an interactive session on the compute node. + +!!! note + + Please note that if you are using GPUs, check whether your script is parallelized. If your script is not parallelized and only depends on GPU, then you don't need to request more cores per node. In that case, do not use `-n` while executing the `interactive` command, as the default option will request 1 CPU per GPU. It's important to keep in mind that using multiple cores on GPU nodes may result in unnecessary CPU hour charges. Additionally, implementing this practice can make service unit accounting significantly easier. \ No newline at end of file diff --git a/docs/Running_jobs/job_limitation.md b/docs/Running_jobs/job_limitation.md new file mode 100644 index 000000000..37b4081e1 --- /dev/null +++ b/docs/Running_jobs/job_limitation.md @@ -0,0 +1,15 @@ +## Overview + +Wulver, like most shared HPC clusters, enforces certain job limitations to ensure fair and efficient resource usage among all users. These limitations are configured through Slurm and can affect the number of jobs, runtime, memory, GPU usage, and priority of execution. Understanding these limits can help you plan better, reduce job failures, and avoid unintentional misuse. + + +## General limitations of job scheduling + +- **Walltime Limits**: `standard`, `low`, and `high` QoS: maximum 72 hours (3 days); `debug` partition: maximum 8 hours. [More info](../Running_jobs/node-memory-config.md/#priority-use-qos) + +- **SUs exhausted**: Once your Service Units are exhausted, you can no longer run your jobs on `standard` or `high` priority but you can still use `low`. + +- **Job preemption**: Jobs running on `low` priority can be preempted by `standard` or `high` priority jobs. + +- **Maintenance**: During the maintenance downtime, logins will be disabled and all the jobs will be held in scheduler. If you submit your job before maintenance with a walltime overlapping the maintenance period then your job will also be held by scheduler. [More info](../faq/faq.md/#maintenance) + diff --git a/docs/Running_jobs/managing-jobs.md b/docs/Running_jobs/managing-jobs.md new file mode 100644 index 000000000..c9b62952c --- /dev/null +++ b/docs/Running_jobs/managing-jobs.md @@ -0,0 +1,234 @@ +## Overview + +Managing and monitoring your jobs effectively helps ensure efficient use of resources and enables quicker debugging when things go wrong. Slurm provides several built-in commands to track job status, usage, and troubleshoot issues. + +SLURM has numerous tools for monitoring jobs. Below are a few to get started. More documentation is available on the [SLURM website](https://slurm.schedmd.com/man_index.html). + +The most common commands are: + +- List all current jobs: `squeue` +- Job deletion: `scancel [job_id]` +- Run a job: `sbatch [submit script]` +- Run a command: `srun
{str(x).strip()}")
+# Apply style to prevent wrapping, hide the index, and convert to HTML
+html_output = df.style.set_properties(**{'white-space': 'nowrap', 'text-align': 'left'}, subset=df.columns[0]).hide(axis='index').to_html()
+print(html_output)
+```
+
+### Priority (Use `--qos`)
+Wulver has three levels of “priority”, utilized under SLURM as Quality of Service (QoS):
+```python exec="on"
+import pandas as pd
+import numpy as np
+df = pd.read_csv('docs/assets/tables/slurm_qos.csv')
+df.replace(np.nan, 'NA', inplace=True)
+df.iloc[:, 0] = df.iloc[:, 0].apply(lambda x: f"{str(x).strip()}")
+# Apply style to prevent wrapping, hide the index, and convert to HTML
+html_output = df.style.set_properties(**{'white-space': 'nowrap', 'text-align': 'left'}, subset=df.columns[0]).hide(axis='index').to_html()
+print(html_output)
+```
+
+## How many cores and memory do I need?
+There is no deterministic method of finding the exact amount of memory needed by a job in advance. A good practice is to overestimate it slightly and then scale down based on previous runs. Significant overestimation, however, can lead to inefficiency of system resources and unnecessary expenditure of CPU time allocations.
+
+We have tool [`seff`](../Running_jobs/managing-jobs.md/#seff-command) in Slurm which you can use to check how much resources your job consumes and based on that re-adjust the configurations.
+
+Understanding where your code is spending time or memory is key to efficient resource usage. Profiling helps you answer questions like:
+
+- Am I using too many CPU cores without benefit?
+- Is my job memory-bound or I/O-bound?
+- Are there inefficient loops, repeated operations, or unused computations?
+
+### Tips for optimization
+- Use multi-threaded or parallel libraries (OpenMP, MPI, NumPy with MKL).
+- Avoid unnecessary data copying or large in-memory objects.
+- Stream large files instead of loading entire datasets into memory.
+- Use job arrays for independent jobs instead of looping in one script.
+
+
+## Be careful about invalid configuration
+Misconfigured job submissions can lead to job failures, wasted compute time, or inefficient resource usage.+Below are some common mistakes and conflicts to watch out for when submitting jobs to SLURM: + +- Asking for more CPUs, memory, or GPUs than any node in the cluster can offer. Job stays in pending state indefinitely with reason like `ReqNodeNotAvail or Resources` +- Mismatch Between CPUs and Tasks. For example: Using `--ntasks=4` and `--cpus-per-task=8` but your script is single-threaded. You're blocking 32 cores but using only 1 effectively — leads to very low CPU efficiency. +- Specifying walltime of more then 3 days is not allowed. +- Submitting to a partition that doesn’t match your job type. For eg. Requesting a GPU with a non-GPU partition: `--partition=standard --gres=gpu:1`. Job will fail immediately or be held forever. + + diff --git a/docs/Running_jobs/ondemand-jobs.md b/docs/Running_jobs/ondemand-jobs.md new file mode 100644 index 000000000..68fdf2468 --- /dev/null +++ b/docs/Running_jobs/ondemand-jobs.md @@ -0,0 +1,20 @@ +# Jobs on OnDemand + +In addition to submitting jobs via batch scripts and interactive sessions, Wulver also supports [Open OnDemand](https://ondemand.njit.edu), a browser-based portal for job management. + +### With OnDemand, users can: + +- Submit and monitor jobs through a graphical interface +- Create and manage [job templates](../OnDemand/6_jobs.md/#jobs) without writing SLURM scripts manually +- Access [interactive applications](../OnDemand/Interactive_Apps/index.md) such as JupyterLab, Matlab, RStudio and many more directly in the browser +- Launch remote desktops on cluster nodes for GUI-based workflows +- Get access to browser based shell [terminal](../OnDemand/3_clusters.md) to access Wulver + +### OnDemand is ideal for: + +- New users who are unfamiliar with Linux and SLURM commands +- Researchers who prefer a graphical interface +- Anyone who wants quick access to interactive HPC applications + +!!! info + See the [**Open OnDemand page**](../OnDemand/index.md) for full documentation. diff --git a/docs/Running_jobs/problems-and-misconceptions.md b/docs/Running_jobs/problems-and-misconceptions.md new file mode 100644 index 000000000..a97dc69c2 --- /dev/null +++ b/docs/Running_jobs/problems-and-misconceptions.md @@ -0,0 +1,86 @@ +# Overview + +New HPC users often assume that requesting more resources (CPUs, GPUs, memory) will automatically make their jobs run faster. In reality, performance depends on how the software is written and configured. Submitting jobs with incorrect resource requests can result in wasted allocations, slower performance, and unnecessary load on shared compute nodes. Below are some common mistakes and their solutions. + +### **Misconception:** “If I allocate more CPUs, my software will automatically use them” + +Many applications are not parallelized by default. Requesting multiple CPUs (--ntasks > 1) will not speed up execution unless your software is explicitly written to take advantage of parallelism (e.g., via MPI, OpenMP, or job arrays). Otherwise, the job may simply run the program multiple times in parallel instead of speeding it up. + +**Example of incorrect job script:** + +```shell +#!/bin/bash -l +#SBATCH --job-name=python +#SBATCH --output=%x.%j.out +#SBATCH --error=%x.%j.err +#SBATCH --partition=general +#SBATCH --ntasks=4 +#SBATCH --qos=standard +#SBATCH --time=30:00 +################################################################################# +module load foss/2024a Python +srun python test.py +``` + +**Problem:** This script launches test.py 4 times since no parallelism is enabled in the code. + +**Solution:** If your code is serial, request only 1 task: `srun -n1 python test.py` or `python test.py` + +To truly leverage multiple CPUs, use parallel programming libraries such as [mpi4py](https://mpi4py.readthedocs.io/en/stable/tutorial.html) and [Parsl](https://parsl.readthedocs.io/en/stable/quickstart.html) + + +### **Misconception:** “My jobs run slower when I request more resources” + +Requesting excessive resources can actually degrade performance. For example, oversubscribing CPUs (assigning more threads than available cores) leads to CPU contention, slowing down computations. + +**Example of problematic job script:** + +```shell +#!/bin/bash -l +#SBATCH -J gmx-test +#SBATCH -o %x.%j.out +#SBATCH -e %x.%j.err +#SBATCH --partition=gpu +#SBATCH --qos=standard +#SBATCH --time 72:00:00 +#SBATCH --nodes=1 +#SBATCH --ntasks-per-node=128 +#SBATCH --gres=gpu:4 +################################################################################# +module purge +module load wulver +module load foss/2025a GROMACS +gmx grompp -f run.mdp -c npt2.gro -r npt2.gro -p topol.top -o run.tpr +srun gmx_mpi mdrun -deffnm run -cpi run.cpt -v -ntomp 2 -pin on -tunepme -dlb yes -noappend +``` + +**Problem:** With `--ntasks-per-node=128` and `-ntomp 2`, the job requests **256** CPUs, but the node only has 128. This overloads the node and slows down execution. + +**Solution:** Match resource requests to the available hardware. For example: + +This job will launch using 64 cores with 2 threads per core. + +```shell +#!/bin/bash -l +#SBATCH -J gmx-test +#SBATCH -o %x.%j.out +#SBATCH -e %x.%j.err +#SBATCH --partition=gpu +#SBATCH --qos=standard +#SBATCH --time=72:00:00 +#SBATCH --nodes=1 +#SBATCH --ntasks-per-node=64 +#SBATCH --cpus-per-task=2 +#SBATCH --gres=gpu:4 +################################################################################# +module purge +module load wulver +module load foss/2025a GROMACS +gmx grompp -f run.mdp -c npt2.gro -r npt2.gro -p topol.top -o run.tpr +srun gmx_mpi mdrun -deffnm run -cpi run.cpt -v -ntomp 2 -pin on -tunepme -dlb yes -noappend +``` + +!!! tips + - Use the checkload command to monitor CPU usage. + - Cancel jobs that overload nodes and adjust scripts accordingly. + - Align --ntasks, --cpus-per-task, and application threading flags (-ntomp, OMP_NUM_THREADS, etc.) with actual node architecture. \ No newline at end of file diff --git a/docs/Running_jobs/service-units.md b/docs/Running_jobs/service-units.md new file mode 100644 index 000000000..405f996f2 --- /dev/null +++ b/docs/Running_jobs/service-units.md @@ -0,0 +1,82 @@ +## Overview + +Service Units (SUs) are the core accounting mechanism used to track and allocate compute usage on Wulver. Each job you run consumes a certain number of SUs based on the resources you request and the duration of your job. + +SUs help us ensure fair usage of the HPC system and monitor consumption across different users, departments, or projects. + +Since resources are limited, each PI's research account is allocated 300,000 Service Units (SUs) per year upon request at no cost. These SUs can be used via the standard [priority](#priority-use-qos) on the SLURM job scheduler. +One SU is defined as 1 CPU hour or 4 GB of RAM per hour. + +!!! tip "Important information on SU" + + * The SU allocation is per PI account, not per individual student. + * The allocation resets each fiscal year. + * Students are expected to use SUs efficiently, as excessive usage may deplete their group's SU balance quickly. + +If a group exhausts its SU allocation early, the PI has the option to purchase additional SUs or leverage higher [priority](#priority-use-qos) queues through investment. For more details, refer to the [Wulver Policies](wulver_policies.md) and [Condo Policies](condo_policies.md). +Check the table below to see how SU will be charged for different partitions. + +```python exec="on" +import pandas as pd +import numpy as np +df = pd.read_csv('docs/assets/tables/SU.csv') +# Replace NaN with 'NA' +df.replace(np.nan, 'NA', inplace=True) +print(df.to_markdown(index=False)) +``` + +!!! warning "Memory request via job scheduler" + + Please note that in the above SU calculation, MAX(CPUs, RAM/4GB) in the `general` and `gpu` partition — this represents the maximum of the number of CPUs requested and the memory requested divided by 4GB. in `bigmem` it's 16 GB. + + * If you do not specify `--mem` in your SLURM job script, the job will be allocated the default 4GB of memory per core (16GB/core in `bigmem`), and you will be charged based on the number of CPUs requested. + * If you do specify `--mem` to request more memory, the SU charge will be based on the maximum of CPU count and memory/4GB (memory/16GB in `bigmem`). + Requesting more memory than the default will result in higher SU charges than if you had only specified CPUs. + + Therefore, please be mindful of the `--mem` setting. Requesting more memory than necessary can significantly increase your SU usage. + +???+ example "Example of SU Charges" + + === "`general` Partition" + | SLURM Directive | SU | Explanation | + |---------------------|:---------:|:---------:| + | 4 CPUs | MAX(4, 4*4G/4G) = 4 | Since no memory requiremnt is specified, SU is charged based on the same number of CPUs | + | 4 CPUs + `--mem=64G` | MAX(4, 64G/4G) = 16 | Since 64G memory is specified, the MAX function the evaluates the maximum of 4 CPUS, and 64G/4G= 16, resulting in a charge of 16 SUs | + | 4 CPUs + `--mem=4G` | MAX(4, 4G/4G) = 4 | MAX function the evaluates the maximum of 4 CPUS, and 4G/4G= 1, resulting in a charge of 4 SUs | + | 4 CPUs + `--mem-per-cpu=8G` | MAX(4, 8G*4/4G) = 8 | MAX function the evaluates the maximum of 4 CPUS, and 8G*4CPUs/4G = 8 , resulting in a charge of 8 SUs | + + === "`bigmem` Partition" + | SLURM Directive | SU | Explanation | + |-----------------------------|:---------------------------:|:----------------------------------------------------------------------------------------------------:| + | 4 CPUs | MAX(4*1.5, 1.5*4*16G/16G) = 6 | On `bigmem` partition the usage factor is 1.5 | + | 4 CPUs + `--mem=64G` | MAX(4*1.5, 1.5*64G/16G) = 6 | Since 64G memory is specified, the MAX function the evaluates the maximum of 4*1.5= 6 SUs, and 1.5*64G/16G= 6 SUs, resulting in a charge of 6 SUs | + | 4 CPUs + `--mem=128G` | MAX(4*1.5, 1.5*128G/16G) = 12 | MAX function the evaluates the maximum of 4*1.5= 6 SUs, and 1.5*128G/16G= 12 SU, resulting in a charge of 12 SUs | + | 4 CPUs + `--mem-per-cpu=8G` | MAX(4*1.5, 1.5*8G*4/16G) = 6 | MAX function the evaluates the maximum of 4*1.5= 6 SUs, and 1.5*8G*4CPUs/16G = 3 SUs , resulting in a charge of 6 SUs | + + === "`gpu` Partition" + | SLURM Directive | SU | Explanation | + |---------------------|:---------:|:---------:| + | 4 CPUs + 10MIG | MAX(4, 4*4G/4G) + 16 * (10G/80G) = 6 | Since no memory requiremnt is specified, SU is charged based on the same number of CPUs and 10G of GPU memory | + | 4 CPUs + 20MIG | MAX(4, 4*4G/4G) + 16 * (20G/80G) = 8 | SU is charged based on the same number of CPUs and 20G of GPU memory | + | 4 CPUs + 40MIG | MAX(4, 4*4G/4G) + 16 * (40G/80G) = 12 | SU is charged based on the same number of CPUs and 40G of GPU memory | + | 4 CPUs + Full GPU | MAX(4, 4*4G/4G) + 16 * (80G/80G) = 20 | SU is charged based on the same number of CPUs and 80G of GPU (A full GPU) memory | + | 4 CPUs + `--mem=64G` + Full GPU | MAX(4, 64G/4G) + 16 * (80G/80G) = 32 | The MAX function evaluates the maximum of 4 SUs (from CPUs), and 64G/4G= 16 SUs (from memory). In addition, 16 SUs are charged from 80G of GPU (A full GPU) memory, bringing the total SU charge to 32 SUs | + | 4 CPUs + `--mem-per-cpu=8G` + Full GPU | MAX(4, 4*8G/4G) + 16 * (80G/80G) = 24 | The MAX function the evaluates the maximum of 4 SUs (from CPUs), and 4*8G/4G= 8 SUs (from memory). In addition, 16 SUs are charged from 80G of GPU (A full GPU) memory, bringing the total SU charge to 24 SUs | + + +### Check Quota + +Users can check their account(s) SU utilization and storage usage via `quota_info UCID` command. +```bash linenums="1" +[ab1234@login01 ~]$ module load wulver +[ab1234@login01 ~]$ quota_info $LOGNAME +Usage for account: xy1234 + SLURM Service Units (CPU Hours): 277557 (300000 Quota) + User ab1234 Usage: 1703 CPU Hours (of 277557 CPU Hours) + PROJECT Storage: 867 GB (of 2048 GB quota) + User ab1234 Usage: 11 GB (No quota) + SCRATCH Storage: 791 GB (of 10240 GB quota) + User ab1234 Usage: 50 GB (No quota) +HOME Storage ab1234 Usage: 0 GB (of 50 GB quota) +``` +Here, `xy1234` represents the UCID of the PI, and "SLURM Service Units (CPU Hours): 277557 (300000 Quota)" indicates that members of the PI group have already utilized 277,557 CPU hours out of the allocated 300,000 SUs, and the user `xy1234` utilized 1703 CPU Hours out of 277,557 CPU Hours. This command also displays the storage usage of directories such as `$HOME`, `/project`, and `/scratch`. Users can view both the group usage and individual usage of each storage. In the given example, the group usage from the 2TB project quota is 867 GB, with the user's usage being 11 GB out of that 867 GB. For more details file system quota, see [Wulver Filesystem](get_started_on_Wulver.md#wulver-filesystems). diff --git a/docs/Services/hpc-services.md b/docs/Services/hpc-services.md new file mode 100644 index 000000000..d0a62fdd4 --- /dev/null +++ b/docs/Services/hpc-services.md @@ -0,0 +1,46 @@ +# HPC Service Catalog + +The NJIT High Performance Computing (HPC) facility provides a wide range of services to support research, teaching, and computation across disciplines. +This catalog outlines the core services available to students, faculty, and researchers. + + +## Cluster Computing +Built by Dell, our computing cluster **Wulver** provides over 197 compute nodes to support computationally intensive workloads across various academic and research domains. +Users can run simulations, data processing, and large-scale modeling efficiently using Wulver’s distributed computing infrastructure. + +Learn more: [Wulver](../clusters/wulver.md) + + +## Research Data Storage +Wulver offers high-performance, multi-tier storage optimized for research. +Each user gets access to dedicated directories for computation, collaboration, and long-term storage. + +- `/home` – Personal files and configurations (50 GB quota). +- `/project` – Primary workspace for research data (2 TB per PI group). +- `/research` – Long-term archival storage, available for purchase by PIs ($100 / TB for five years). + +Learn more: [File Systems and Storage on Wulver](../clusters/Wulver_filesystems.md) + + +## Education +The HPC facility supports **course integration and instructional use**, offering dedicated partitions and allocations for coursework that require high-performance computing environments. +These resources help students gain hands-on experience with parallel computing and data-intensive applications. + +Learn more: [Resources for Coursework](../Courses/index.md) + +## Scientific Software Development +The HPC team provides deep expertise in **developing, installing, and optimizing scientific software** for Wulver. +We ensure that applications are configured for maximum performance in the cluster environment. We provide a wide range of software for research purposes, and you may also request the installation of additional software, subject to our guidelines and approval. + +Learn more: [Available Software List](../Software/index.md) + + +## HPC Facilitation Service +Our facilitation team empowers users to perform essential research computing projects through **training, webinars, one-on-one consultation, and effective user support**. We also have in person office hours held by our student interns. + +Learn more: [Contact the HPC Facilitation Team](../about/contact.md) + + +!!! info "Reach out to us" + For any inquiries or assistance with HPC resources, please reach out to us through our email: hpc@njit.edu + diff --git a/docs/Software/.pages b/docs/Software/.pages index e69de29bb..f172f9a90 100644 --- a/docs/Software/.pages +++ b/docs/Software/.pages @@ -0,0 +1,2 @@ +title: Software +collapse: true \ No newline at end of file diff --git a/docs/Software/CFD/ansys.md b/docs/Software/CFD/ansys.md index f35c69c3e..859520363 100644 --- a/docs/Software/CFD/ansys.md +++ b/docs/Software/CFD/ansys.md @@ -12,17 +12,7 @@ The software suite is known for its high level of accuracy and versatility, and ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "ANSYS"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "ANSYS"') print(soft.to_markdown(index=False)) ``` @@ -41,6 +31,6 @@ Please [download](https://njit.instructure.com/courses/8519/assignments/128626) !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/CFD/comsol.md b/docs/Software/CFD/comsol.md index 390628431..cf8e8d1cf 100644 --- a/docs/Software/CFD/comsol.md +++ b/docs/Software/CFD/comsol.md @@ -14,20 +14,11 @@ COMSOL is widely used in engineering and science fields, such as mechanical engi ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "COMSOL"') print(soft.to_markdown(index=False)) ``` -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "COMSOL" | Software == "comsol"') - print(soft.to_markdown(index=False)) - ``` ## Application Information, Documentation @@ -41,6 +32,6 @@ COMSOL is widely used in engineering and science fields, such as mechanical engi !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/CFD/fluent.md b/docs/Software/CFD/fluent.md index 4c2a493df..bcae51079 100644 --- a/docs/Software/CFD/fluent.md +++ b/docs/Software/CFD/fluent.md @@ -10,17 +10,7 @@ title: FLUENT ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "fluent" | Software == "ANSYS"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "fluent" | Software == "ANSYS"') print(soft.to_markdown(index=False)) ``` @@ -59,8 +49,11 @@ If you are running transient problem and want to save the data at particular tim (print-case-timer) parallel/timer/usage ``` -In the above `Journal` script, the full name of case file (`tube_vof.cas.h5`) is mentioned. You need to modify based on the case file based on the problem. The `solve/dual-time-iterate` specifies the end flow time and number of iterations. In the above example, `20` is the end flow time while the maximum number of iterations are `50`. The "dual-time" approach allows for a larger time step size by introducing an additional iteration loop within each time step. Users can select different approach based on their problems and need to modify it accordingly. +In the above `Journal` script, the full name of case file (`tube_vof.cas.h5`) is mentioned. You need to modify based on the case file based on the problem. +The `solve/dual-time-iterate` specifies the end flow time and number of iterations. In the above example, `20` is the end flow time while the maximum number of iterations are `50`. The "dual-time" approach allows for a larger time step size by introducing an additional iteration loop within each time step. Users can select different approach based on their problems and need to modify it accordingly. + ??? example "Sample Batch Script to Run FLUENT : fluent.submit.sh" @@ -99,38 +92,7 @@ For more details on journal commands, see the Fluent text user interface (TUI) c fluent 3ddp -affinity=off -ssh -t$SLURM_NTASKS -pib -mpi=intel -cnf="$machines" -g -i journal.JOU ``` - === "Lochness" - - ```slurm - #!/bin/bash -l - #SBATCH --job-name=fluent - #SBATCH --output=%x.%j.out # i%x.%j expands to slurm JobName.JobID - #SBATCH --error=%x.%j.err # prints the error message - #SBATCH --ntasks=8 - # Use "sinfo" to see what partitions are available to you - #SBATCH --partition=public - - # Memory required; lower amount gets scheduling priority - #SBATCH --mem-per-cpu=5G - - # Time required in d-hh:mm:ss format; lower time gets scheduling priority - #SBATCH --time=5-24:59:00 - - # Purge and load the correct modules - module purge > /dev/null 2>&1 - module load ANSYS - - # Run the mpi program - - machines=hosts.$SLURM_JOB_ID - touch $machines - for node in `scontrol show hostnames` - do - echo "$node" >> $machines - done - - fluent 3ddp -affinity=off -ssh -t$SLURM_NTASKS -pib -mpi=intel -cnf="$machines" -g -i journal.JOU - ``` + Submit the job using `sbatch fluent.submit.sh` command. @@ -142,6 +104,6 @@ Submit the job using `sbatch fluent.submit.sh` command. !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/CFD/openfoam.md b/docs/Software/CFD/openfoam.md index b1060a77c..dae254c10 100644 --- a/docs/Software/CFD/openfoam.md +++ b/docs/Software/CFD/openfoam.md @@ -14,17 +14,7 @@ The software is widely used in academia, research, and industry, and is known fo ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "OpenFOAM"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "OpenFOAM"') print(soft.to_markdown(index=False)) ``` @@ -58,7 +48,7 @@ OpenFOAM can be used for both serial and parallel jobs. To run OpenFOAM in paral ################################################ module purge module load wulver # Load slurm, easybuild - module load foss/2021b OpenFOAM + module load foss/2024a OpenFOAM ################################################ # # Source OpenFOAM bashrc @@ -83,53 +73,12 @@ OpenFOAM can be used for both serial and parallel jobs. To run OpenFOAM in paral reconstructPar ``` - === "Lochness" - - ```slurm - #!/bin/bash -l - #SBATCH --job-name=openfoam_parallel - #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID - #SBATCH --partition=public - #SBATCH --nodes=1 - #SBATCH --ntasks-per-node=16 - #SBATCH --mem-per-cpu=10G # Adjust as necessary - #SBATCH --time=00:01:00 # D-HH:MM:SS - ################################################ - # - # Purge and load modules needed for run - # - ################################################ - module purge - module load foss/2021b OpenFOAM - ################################################ - # - # Source OpenFOAM bashrc - # The modulefile doesn't do this - # - ################################################ - source $FOAM_BASH - ################################################ - # - # cd into cavity directory and run blockMesh and - # icoFoam. Note: this is running on one node and - # using all 32 cores on the node - # - ################################################ - cd cavity - blockMesh - decomposePar -force - srun icoFoam -parallel - reconstructPar - ``` !!! note === "Wulver" You can copy the tutorial `cavity` mentioned in the above job script from the `/apps/easybuild/examples/openFoam/parallel` directory. - === "Lochness" - - You can copy the tutorial `cavity` mentioned in the above job script from the `/opt/site/examples/openFoam/parallel` directory. To run OpenFOAM in serial, the following job script can be used. @@ -179,44 +128,6 @@ To run OpenFOAM in serial, the following job script can be used. icoFoam ``` - === "Lochness" - - ```slurm - #!/bin/bash -l - #SBATCH --job-name=openfoam_serial - #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID - #SBATCH --partition=public - #SBATCH --nodes=1 - #SBATCH --ntasks-per-node=1 - #SBATCH --mem-per-cpu=10G # Adjust as necessary - #SBATCH --time=00:01:00 # D-HH:MM:SS - ################################################ - # - # Purge and load modules needed for run - # - ################################################ - module purge - module load foss/2021b OpenFOAM - ################################################ - # - # Source OpenFOAM bashrc - # The modulefile doesn't do this - # - ################################################ - source $FOAM_BASH - ################################################ - # - # copy into cavity directory from /opt/site/examples/openFoam/parallel and run blockMesh and - # icoFoam. Note: this is running on one node and - # using all 32 cores on the node - # - ################################################ - cp -r /opt/site/examples/openFoam/parallel/cavity /path/to/destination - # /path/to/destination is destination path where user wants to copy the cavity directory - cd cavity - blockMesh - icoFoam - ``` Submit the job script using the sbatch command: `sbatch openfoam_parallel.submit.sh` or `sbatch openfoam_serial.submit.sh`. ## Building OpenFOAM from source @@ -224,8 +135,11 @@ Sometimes, users need to create a new solver or modify the existing solver by ad ```bash - # This is to build a completly self contained OpenFOAM using MPICH mpi. Everything from GCC on up will be built. - + # This is to build a completely self contained OpenFOAM using MPICH mpi. Everything from GCC on up will be built. + + # start an interactive session with compute node. Replace "PI_UCID" with the UCID of PI. Modify the other parameters if required. + srun --partition=general --nodes=1 --ntasks-per-node=16 --mem-per-cpu=2G --account=PI_UCID --qos=standard --time=2:00:00 --pty bash + # purge all loaded modules module purge # Download the latest version of OpenFOAM, visit https://develop.openfoam.com/Development/openfoam/-/blob/master/doc/Build.md for details @@ -302,7 +216,7 @@ Sometimes, users need to create a new solver or modify the existing solver by ad !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/IDE/VSCode.md b/docs/Software/IDE/VSCode.md index 447be2da7..f0c19ac7f 100644 --- a/docs/Software/IDE/VSCode.md +++ b/docs/Software/IDE/VSCode.md @@ -18,86 +18,48 @@ The documentation of VS Code is available at [VS Code documentation](https://cod Use the following slurm script and submit the job script using `sbatch vs-code.submit.sh` command. ??? example "Batch Script to use VS Code : vs-code.submit.sh" - - === "Wulver" - - ```slurm - #!/bin/bash -l - #SBATCH --job-name=vs-code - #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID - #SBATCH --error=%x.%j.err # prints the error message - #SBATCH --partition=general - #SBATCH --nodes=1 - #SBATCH --ntasks-per-node=32 - #SBATCH --mem-per-cpu=4000M # Maximum allowable mempry per CPU 4G - #SBATCH --qos=standard - #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI - #SBATCH --time=71:59:59 # D-HH:MM:SS - - set -e - - module purge - module load wulver # load slurn, easybuild - # add any required module loads here, e.g. a specific Python - - CLI_PATH="${HOME}/vscode_cli" - - # Install the VS Code CLI command if it doesn't exist - if [[ ! -e ${CLI_PATH}/code ]]; then - echo "Downloading and installing the VS Code CLI command" - mkdir -p "${HOME}/vscode_cli" - pushd "${HOME}/vscode_cli" - # Process from: https://code.visualstudio.com/docs/remote/tunnels#_using-the-code-cli - curl -Lk 'https://code.visualstudio.com/sha/download?build=stable&os=cli-alpine-x64' --output vscode_cli.tar.gz - # unpack the code binary file - tar -xf vscode_cli.tar.gz - # clean-up - rm vscode_cli.tar.gz - popd - fi + ```slurm + #!/bin/bash -l + #SBATCH --job-name=vs-code + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.out # prints the error message + #SBATCH --partition=general + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=32 + #SBATCH --mem-per-cpu=4000M # Maximum allowable mempry per CPU 4G + #SBATCH --qos=standard + #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI + #SBATCH --time=71:59:59 # D-HH:MM:SS + + set -e + + module purge + module load wulver # load slurn, easybuild - # run the code tunnel command and accept the licence - ${CLI_PATH}/code tunnel --accept-server-license-terms - ``` + # add any required module loads here, e.g. a specific Python + + CLI_PATH="${HOME}/vscode_cli" + + # Install the VS Code CLI command if it doesn't exist + if [[ ! -e ${CLI_PATH}/code ]]; then + echo "Downloading and installing the VS Code CLI command" + mkdir -p "${HOME}/vscode_cli" + pushd "${HOME}/vscode_cli" + # Process from: https://code.visualstudio.com/docs/remote/tunnels#_using-the-code-cli + curl -Lk 'https://code.visualstudio.com/sha/download?build=stable&os=cli-alpine-x64' --output vscode_cli.tar.gz + # unpack the code binary file + tar -xf vscode_cli.tar.gz + # clean-up + rm vscode_cli.tar.gz + popd + fi + + # run the code tunnel command and accept the licence + ${CLI_PATH}/code tunnel --accept-server-license-terms + ``` - === "Lochness" - - ```slurm - #!/bin/bash -l - #SBATCH --job-name=vs-code - #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID - #SBATCH --partition=datasci - #SBATCH --nodes=1 - #SBATCH --ntasks-per-node=8 - #SBATCH --time=24:59:00 # D-HH:MM:SS - #SBATCH --mem-per-cpu=4G - - set -e - - module purge - - # add any required module loads here, e.g. a specific Python - - CLI_PATH="${HOME}/vscode_cli" - - # Install the VS Code CLI command if it doesn't exist - if [[ ! -e ${CLI_PATH}/code ]]; then - echo "Downloading and installing the VS Code CLI command" - mkdir -p "${HOME}/vscode_cli" - pushd "${HOME}/vscode_cli" - # Process from: https://code.visualstudio.com/docs/remote/tunnels#_using-the-code-cli - curl -Lk 'https://code.visualstudio.com/sha/download?build=stable&os=cli-alpine-x64' --output vscode_cli.tar.gz - # unpack the code binary file - tar -xf vscode_cli.tar.gz - # clean-up - rm vscode_cli.tar.gz - popd - fi - # run the code tunnel command and accept the licence - ${CLI_PATH}/code tunnel --accept-server-license-terms - ``` Once you submit the job, you will see an output file with `.out` extension. Once you open the file, you will see the following ``` * @@ -117,7 +79,7 @@ Once you submit the job, you will see an output file with `.out` extension. Once To grant access to the server, please log into https://github.com/login/device and use code XXXX-XXXX ``` -You need to have the [GitHub](https://wwww.github.com) account, please open the GitHub profile and use the code printed in the output file. Once you authorize GitHub, you will see the following in the output file +You need to have the [GitHub](https://www.github.com) account, please open the GitHub profile and use the code printed in the output file. Once you authorize GitHub, you will see the following in the output file ``` Open this link in your browser https://vscode.dev/tunnel/nodeXXX @@ -134,6 +96,6 @@ Now copy and paste this link in your browser and VS Code is ready to use. !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/math/MATLAB/img/ClickOnAddons.png b/docs/Software/MATLAB/img/ClickOnAddons.png similarity index 100% rename from docs/Software/math/MATLAB/img/ClickOnAddons.png rename to docs/Software/MATLAB/img/ClickOnAddons.png diff --git a/docs/Software/math/MATLAB/img/ClickOnInstall.png b/docs/Software/MATLAB/img/ClickOnInstall.png similarity index 100% rename from docs/Software/math/MATLAB/img/ClickOnInstall.png rename to docs/Software/MATLAB/img/ClickOnInstall.png diff --git a/docs/Software/MATLAB/img/FetchOutputs.png b/docs/Software/MATLAB/img/FetchOutputs.png new file mode 100644 index 000000000..aeec77029 Binary files /dev/null and b/docs/Software/MATLAB/img/FetchOutputs.png differ diff --git a/docs/Software/math/MATLAB/img/GenericProfile1.png b/docs/Software/MATLAB/img/GenericProfile1.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile1.png rename to docs/Software/MATLAB/img/GenericProfile1.png diff --git a/docs/Software/math/MATLAB/img/GenericProfile10.png b/docs/Software/MATLAB/img/GenericProfile10.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile10.png rename to docs/Software/MATLAB/img/GenericProfile10.png diff --git a/docs/Software/math/MATLAB/img/GenericProfile11.png b/docs/Software/MATLAB/img/GenericProfile11.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile11.png rename to docs/Software/MATLAB/img/GenericProfile11.png diff --git a/docs/Software/math/MATLAB/img/GenericProfile2.png b/docs/Software/MATLAB/img/GenericProfile2.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile2.png rename to docs/Software/MATLAB/img/GenericProfile2.png diff --git a/docs/Software/math/MATLAB/img/GenericProfile3.png b/docs/Software/MATLAB/img/GenericProfile3.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile3.png rename to docs/Software/MATLAB/img/GenericProfile3.png diff --git a/docs/Software/math/MATLAB/img/GenericProfile4.png b/docs/Software/MATLAB/img/GenericProfile4.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile4.png rename to docs/Software/MATLAB/img/GenericProfile4.png diff --git a/docs/Software/MATLAB/img/GenericProfile5.png b/docs/Software/MATLAB/img/GenericProfile5.png new file mode 100644 index 000000000..bd0deac8f Binary files /dev/null and b/docs/Software/MATLAB/img/GenericProfile5.png differ diff --git a/docs/Software/math/MATLAB/img/GenericProfile6.png b/docs/Software/MATLAB/img/GenericProfile6.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile6.png rename to docs/Software/MATLAB/img/GenericProfile6.png diff --git a/docs/Software/MATLAB/img/GenericProfile7.png b/docs/Software/MATLAB/img/GenericProfile7.png new file mode 100644 index 000000000..58c6698b5 Binary files /dev/null and b/docs/Software/MATLAB/img/GenericProfile7.png differ diff --git a/docs/Software/math/MATLAB/img/GenericProfile8.png b/docs/Software/MATLAB/img/GenericProfile8.png similarity index 100% rename from docs/Software/math/MATLAB/img/GenericProfile8.png rename to docs/Software/MATLAB/img/GenericProfile8.png diff --git a/docs/Software/MATLAB/img/GenericProfile9.png b/docs/Software/MATLAB/img/GenericProfile9.png new file mode 100644 index 000000000..ce67545d4 Binary files /dev/null and b/docs/Software/MATLAB/img/GenericProfile9.png differ diff --git a/docs/Software/math/MATLAB/img/InstallationComplete.png b/docs/Software/MATLAB/img/InstallationComplete.png similarity index 100% rename from docs/Software/math/MATLAB/img/InstallationComplete.png rename to docs/Software/MATLAB/img/InstallationComplete.png diff --git a/docs/Software/math/MATLAB/img/SlurmAddOn.png b/docs/Software/MATLAB/img/SlurmAddOn.png similarity index 100% rename from docs/Software/math/MATLAB/img/SlurmAddOn.png rename to docs/Software/MATLAB/img/SlurmAddOn.png diff --git a/docs/Software/math/MATLAB/img/matlab_installation_1.png b/docs/Software/MATLAB/img/matlab_installation_1.png similarity index 100% rename from docs/Software/math/MATLAB/img/matlab_installation_1.png rename to docs/Software/MATLAB/img/matlab_installation_1.png diff --git a/docs/Software/math/MATLAB/img/matlab_installation_2.png b/docs/Software/MATLAB/img/matlab_installation_2.png similarity index 100% rename from docs/Software/math/MATLAB/img/matlab_installation_2.png rename to docs/Software/MATLAB/img/matlab_installation_2.png diff --git a/docs/Software/math/MATLAB/img/matlab_installation_3.png b/docs/Software/MATLAB/img/matlab_installation_3.png similarity index 100% rename from docs/Software/math/MATLAB/img/matlab_installation_3.png rename to docs/Software/MATLAB/img/matlab_installation_3.png diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_1.png b/docs/Software/MATLAB/img/matlab_slurm_profile_1.png new file mode 100644 index 000000000..2184144a4 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_1.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_2.png b/docs/Software/MATLAB/img/matlab_slurm_profile_2.png new file mode 100644 index 000000000..cd605fb57 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_2.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_3.png b/docs/Software/MATLAB/img/matlab_slurm_profile_3.png new file mode 100644 index 000000000..ac06f60e1 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_3.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings.png new file mode 100644 index 000000000..74e2055be Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_1.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_1.png new file mode 100644 index 000000000..2c7f7ab89 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_1.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_2.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_2.png new file mode 100644 index 000000000..bf2b4db49 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_2.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_3.PNG b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_3.PNG new file mode 100644 index 000000000..93834007c Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_3.PNG differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_4.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_4.png new file mode 100644 index 000000000..9e77b5bec Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_4.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_5.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_5.png new file mode 100644 index 000000000..4205b284b Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_5.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_6.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_6.png new file mode 100644 index 000000000..dde8e1960 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_6.png differ diff --git a/docs/Software/MATLAB/img/matlab_slurm_profile_settings_7.png b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_7.png new file mode 100644 index 000000000..94ccce351 Binary files /dev/null and b/docs/Software/MATLAB/img/matlab_slurm_profile_settings_7.png differ diff --git a/docs/Software/MATLAB/img/md8jfxI.jpeg b/docs/Software/MATLAB/img/md8jfxI.jpeg new file mode 100644 index 000000000..dbacc3b11 Binary files /dev/null and b/docs/Software/MATLAB/img/md8jfxI.jpeg differ diff --git a/docs/Software/math/MATLAB/index.md b/docs/Software/MATLAB/index.md similarity index 71% rename from docs/Software/math/MATLAB/index.md rename to docs/Software/MATLAB/index.md index cc516a02c..d7ff0e9c6 100644 --- a/docs/Software/math/MATLAB/index.md +++ b/docs/Software/MATLAB/index.md @@ -9,23 +9,13 @@ MATLAB (matrix laboratory) is a multi-paradigm numerical computing environment a ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "MATLAB" | Software == "matlab"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "MATLAB" | Software == "matlab"') print(soft.to_markdown(index=False)) ``` ## Application Information, Documentation -The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathworks.com/support/learn-with-matlab-tutorials.html) +The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathworks.com/support/learn-with-matlab-tutorials.html). ## Using MATLAB @@ -40,7 +30,7 @@ The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathwo #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID #SBATCH --error=%x.%j.err # prints the error message #SBATCH --partition=general - #SBATCH -nodes=1 + #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --mem-per-cpu=4000M # Maximum allowable mempry per CPU 4G #SBATCH --qos=standard @@ -52,25 +42,7 @@ The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathwo module load wulver # Load the slurm, easybuild module load MATLAB - matlab --nodisplay --nosplash -r test - - ``` - - === "Lochness" - - ```slurm - #!/bin/bash - #SBATCH -J test_matlab - #SBATCH --partition=public - #SBATCH -nodes=1 - #SBATCH --ntasks-per-node=1 - #SBATCH - t 30:00 - - # Load matlab module - module purge - module load MATLAB/2022a - - matlab --nodisplay --nosplash -r test + matlab -nodisplay -nosplash -r test ``` @@ -78,7 +50,7 @@ The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathwo ```matlab A = [ 1 2; 3 4] - A.**2 + A.^2 ``` ### Parallel Job @@ -91,8 +63,10 @@ The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathwo ```slurm #!/bin/bash #SBATCH -J test_matlab + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err # prints the error message #SBATCH --partition=general - #SBATCH -nodes=1 + #SBATCH --nodes=1 #SBATCH --ntasks-per-node=32 #SBATCH --mem-per-cpu=4000M # Maximum allowable mempry per CPU 4G #SBATCH --qos=standard @@ -105,26 +79,10 @@ The documentation of MATLAB is available at [MATLAB Tutorial](https://www.mathwo module load MATLAB # Run matlab - matlab -nodisplay --nosplash -r for_loop.m + matlab -nodisplay -nosplash -r 'cd('/path/to/for_loop.m');for_loop; quit' ``` + Replace `cd('/path/to/for_loop.m')` with the actual path of the matlab script. You don't need to use `cd('/path/to/for_loop.m')` if the Matlab script and job script are in the same directory. In that case, use `matlab -nodisplay -nosplash -r 'for_loop; quit' ` - === "Lochness" - - ```slurm - #!/bin/bash - #SBATCH -J test_matlab - #SBATCH --partition=public - #SBATCH -nodes=1 - #SBATCH --ntasks-per-node=32 - #SBATCH - t 30:00 - - # Load matlab module - module purge - module load MATLAB/2022a - - # Run matlab - matlab -nodisplay --nosplash -r for_loop.m - ``` ??? example "Sample Parallel MATLAB script: for_loop.m" diff --git a/docs/Software/MATLAB/matlab_local.md b/docs/Software/MATLAB/matlab_local.md new file mode 100644 index 000000000..5827d5cc6 --- /dev/null +++ b/docs/Software/MATLAB/matlab_local.md @@ -0,0 +1,281 @@ +# Use MATLAB on NJIT HPC + +!!! tip + + Since MFA is enabled, the instructions for running MATLAB via HPC resources have been modified. If you already installed MATLAB on the local machine, skip to [Setup Slurm profile to run MATLAB on Wulver](matlab_local.md#setup-slurm-profile-to-run-matlab-on-wulver). + +## Installation steps of MATLAB on local machine +* Go to [Mathworks Download](https://www.mathworks.com/downloads/) and register with your NJIT email address. +* Select the [MATLAB version](../#availability) installed on Wulver. +* User needs to select the correct installer based on the OS (Mac or Windows). +* Run the installer. + + { width=50% height=50%} + { width=50% height=50%} + +* Make sure to check **Parallel Computing Toolbox** option. + + {width=50% height=50%} + +* Continue by selecting **Next** and MATLAB will be installed on your computer. + +## Setup Slurm profile to run MATLAB on Wulver +* Open MATLAB --> select Create and Manage Clusters. + +{ width=50% height 50%} + +* A new dialogue box will open and under the Add Cluster Profile, select Slurm. + +{ width=50% height 50%} + +{ width=50% height 50%} + +* This will open a Slurm cluster Profile and select the edit option to modify the parameters + +{ width=50% height 50%} + +* Modify the following parameters as mentioned in the screenshot + + + +a. `Description` - Set the name as `Wulver` + +b. `JobStorageLocation` - No Change + +c. `NumWorkers` - 512 + +d. `NumThreads` - No Change + +e. `ClusterMatlabRoot` - Use `module av MATLAB` command first. + +```bash + login-1-45 ~ >: module av MATLAB + ------------------------------------/apps/easybuild/modules/all/Core--------------------------------------------------------- + MATLAB/2024a + +Use "module spider" to find all possible modules and extensions. +Use "module keyword key1 key2 ..." to search for all possible modules matching any of the "keys". +``` +This will show you the list of MATLAB versions installed on Wulver. Next, use `module show MATLAB/2024a` to check MATLAB installation path. + +```bash + login-1-45 ~ >: module show MATLAB/2024a +--------------------------------------------------------------------------------------------------------------------------------- + /apps/easybuild/modules/all/Core/MATLAB/2024a.lua: +--------------------------------------------------------------------------------------------------------------------------------- +help([[ +Description +=========== +The MATLAB Parallel Server Toolbox. + + +More information +================ + - Homepage: https://www.mathworks.com/help/matlab/matlab-engine-for-python.html +]]) +whatis("Description: The MATLAB Parallel Server Toolbox.") +whatis("Homepage: https://www.mathworks.com/help/matlab/matlab-engine-for-python.html") +whatis("URL: https://www.mathworks.com/help/matlab/matlab-engine-for-python.html") +conflict("MATLAB") +prepend_path("CMAKE_PREFIX_PATH","/apps/easybuild/software/MATLAB/2024a") +prepend_path("PATH","/apps/easybuild/software/MATLAB/2024a/bin") +setenv("EBROOTMATLAB","/apps/easybuild/software/MATLAB/2024a") +setenv("EBVERSIONMATLAB","2024a") +setenv("EBDEVELMATLAB","/apps/easybuild/software/MATLAB/R2023a/easybuild/Core-MATLAB-2023a-easybuild-devel") +prepend_path("PATH","/apps/easybuild/software/MATLAB/2024a/toolbox/parallel/bin") +prepend_path("PATH","/apps/easybuild/software/MATLAB/2024a") +prepend_path("LD_LIBRARY_PATH","/apps/easybuild/software/MATLAB/2024a/runtime/glnxa64") +prepend_path("LD_LIBRARY_PATH","/apps/easybuild/software/MATLAB/2024a/bin/glnxa64") +prepend_path("LD_LIBRARY_PATH","/apps/easybuild/software/MATLAB/2024a/sys/os/glnxa64") +setenv("_JAVA_OPTIONS","-Xmx2048m") +``` +The MATLAB installation path is defined by the `EBROOTMATLAB` environment variable, which, in the above example, is set to `/apps/easybuild/software/MATLAB/2024a`. + +f. `RequireOnlineLicensing` - false + +g. `AdditionalProperties` - Select add and add the following as mentioned in the table. + + + + + +| Name | Value | Type | +|----------------------------|:-----------------:|:-------:| +| `ClusterHost` | `wulver.njit.edu` | String | +| `AuthenticationMode` | Multifactor | String | +| `UseUniqueSubfolders` | True | Logical | +| `UseIdentityFile` | False | Logical | +| `RemoteJobStorageLocation` | `$PATH` | String | +| `user` | `$UCID` | String | + +Replace `$PATH` with the actual path of Wulver where you want to save the output file. Make sure to use `/project` directory for remote job storage as `$HOME` has fixed quota of 50GB and cannot be increased. See [Wulver Filesystems](Wulver_filesystems.md) for details. Replace `$UCID` with the NJIT UCID. + + +## Submitting a Serial Job +This section will demonstrate how to create a cluster object and submit a simple job to the cluster. The job will run the 'hostname' command on the node assigned to the job. The output will indicate clearly that the job ran on the cluster and not on the local computer. + +The hostname.m file used in this demonstration can be downloaded [here](https://www.mathworks.com/matlabcentral/fileexchange/24096-hostname-m). + +``` + >> c=parcluster +``` + +Certain arguments need to be passed to SLURM in order for the job to run properly. Here we will set values for partition, and time. In the Matlab window enter: +``` + >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=general --qos=standard --account=$PI_UCID --time=2-00:00:00'] +``` +Replace `$PI_UCID` with the UCID of PI. Check the [Batch Jobs](batch-jobs.md) for other SLURM parameters. +To make this persistent between Matlab sessions these arguments need to be saved to the profile. In the Matlab window enter: +``` + >> c.saveProfile +``` + +We will now submit the hostname.m function to the cluster. In the Matlab window enter the following: +``` +>> j=c.batch(@hostname, 1, {}, 'AutoAddClientPath', false); +``` + +* `@`: Submitting a function. + +* `1`: The number of output arguments from the evaluated function. + +* `{}`: Cell array of input arguments to the function. In this case empty. + +* `'AutoAddClientPath', false`: The client path is not available on the cluster. + + +When the job is submitted, you will be prompted for your password. + +To wait for the job to finish, enter the following in the Matlab window: +``` + >>j.wait +``` +Finally, to get the results: +``` + >>fetchOutputs(j) +``` + +### Submitting a Parallel Function +The `Job Monitor` is a convenient way to monitor jobs submitted to the cluster. In the Matlab window select `Parallel` and then `Monitor Jobs`. + +For more information see the Mathworks page: [Job Monitor](https://www.mathworks.com/help/parallel-computing/job-monitor.html). + +Here we will submit a simple function using a "parfor" loop. The code for this example is as follows: +``` +function t = parallel_example + +t0 = tic; +parfor idx = 1:16 + A(idx) = idx; + pause (2) +end + +t=toc(t0); +``` +To submit this job: +``` + >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=general --qos=standard --account=$PI_UCID --ntasks=8 --time=2-00:00:00'] + >> c.saveProfile + >> j=c.batch(@parallel_example, 1, {}, 'AutoAddClientPath', false, 'Pool', 7) +``` +Since this is a parallel job a 'Pool' must be started. The actual number of tasks started will be one more than requested in the pool. In this case, the batch command calls for a pool of seven. Eight tasks will be started on the cluster. + +The job takes a few minutes to run and the state of the job changes to "finished." + +Once again to get the results enter: +``` + >> fetchOutputs(j) +``` +As can be seen the parfor loop was completed in 6.7591 seconds. + + { width=70% height 70%} + +## Submitting a Script Requiring a GPU +In this section we will submit a matlab script using a GPU. The results will be written to the job diary. The code for this example is as follows: +``` +% MATLAB script that defines a random matrix and does FFT +% +% The first FFT is without a GPU +% The second is with the GPU +% +% MATLAB knows to use the GPU the second time because it +% is passed a type gpuArray as an argument to FFT +% We do the FFT a bunch of times to make using the GPU worth it, +% or else it spends more time offloading to the GPU +% than performning the calculation +% +% This example is meant to provide a general understanding +% of MATLAB GPU usage +% Meaningful performance measurements depend on many factors +% beyond the scope of this example +% Downloaded from https://projects.ncsu.edu/hpc/Software/examples/matlab/gpu/gpu_m + +% Define a matrix +A1 = rand(3000,3000); + +% Just use the compute node, no GPU +tic; +% Do 1000 FFT's +for i = 1:1000 + B2 = fft(A1); +end +time1 = toc; +fprintf('%s\n',"Time to run FFT on the node:") +disp(time1); + +% Use GPU +tic; +A2 = gpuArray(A1); +% Do 1000 FFT's +for i = 1:1000 + % MALAB knows to use GPU FFT because A2 is defined by gpuArray + B2 = fft(A2); +end +time2 = toc; +fprintf('%s\n',"Time to run FFT on the GPU:") +disp(time2); + +% Will be greater than 1 if GPU is faster +speedup = time1/time2 +``` +We will need to change the partition to `gpu` to request a gpu. In the Matlab window enter: +``` + >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=gpu --qos=standard --account=PI_UCID --gres=gpu:1 --mem-per-cpu=4G --time=2-00:00:00'] +``` + { width=70% height 70%} + +Submit the job as before. Since a script is submitted as opposed to a function, only the name of the script is included in the batch command. Do not include the `@` symbol. In a script there are no inputs or ouptuts. +``` + >> j=c.batch('gpu', 'AutoAddClientPath', false) +``` + +To get the result: +``` + >> j.diary +``` + +## Load and Plot Results from A Job +In this section we will run a job on the cluster and then load and plot the results in the local Matlab workspace. The code for this example is as follows: +``` +n=100; +disp("n = " + n); +A = gallery('poisson',n-2); +b = convn(([1,zeros(1,n-2),1]'|[1,zeros(1,n-1)]), 0.5*ones(3,3),'valid')'; +x = reshape(A\b(:),n-2,n-2)';% +``` +As before submit the job: +``` + >> j=c.batch('plot_demo', 'AutoAddClientPath', false); +``` + +To load 'x' into the local Matlab workspace: +``` + >> load(j,'x') +``` + + Finally, plot the results: +``` + >> plot(x) +``` + + \ No newline at end of file diff --git a/docs/Software/Molecular_Dynamics/gromacs.md b/docs/Software/Molecular_Dynamics/gromacs.md new file mode 100644 index 000000000..78bb47cc6 --- /dev/null +++ b/docs/Software/Molecular_Dynamics/gromacs.md @@ -0,0 +1,133 @@ +--- +title: GROMACS +--- + +[GROMACS](https://www.gromacs.org) is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. + +It is primarily designed for biochemical molecules like proteins, lipids, and nucleic acids that have a lot of complicated bonded interactions, but since GROMACS is extremely fast at calculating the nonbonded interactions (that usually dominate simulations) many groups are also using it for research on non-biological systems, e.g. polymers. + +## Availability + +=== "Wulver" + + ```python exec="on" + import pandas as pd + + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') + soft = df.query('Software == "GROMACS"') + print(soft.to_markdown(index=False)) + ``` + +## Application Information, Documentation +The documentation of GROMACS is available at [GROMACS Manual](https://manual.gromacs.org/current/index.html), where you can find the tutorials in topologies, input file format, setting parameters, etc. + +## Using GROMACS +GROMACS can be used on CPU or GPU. When using GROMACS with GPUs (Graphics Processing Units), the calculations can be significantly accelerated, allowing for faster simulations. You can use GROMACS with GPU acceleration, but you need to use GPU nodes on our cluster. + +??? example "Sample Batch Script to Run GROMACS" + + === "GPU" + + ```slurm + #!/bin/bash -l + # NOTE the -l (login) flag! + #SBATCH -J gmx2023 + #SBATCH -o test.%x.%j.out + #SBATCH -e test.%x.%j.err + #SBATCH --mail-type=ALL + #SBATCH --partition=gpu + #SBATCH --qos=standard + #SBATCH --time 72:00:00 # Max 3 days + #SBATCH --nodes=2 + #SBATCH --ntasks-per-node=2 + #SBATCH --gpus-per-node=2 + #SBATCH --account=$PI_ucid # Replace PI_ucid with the UCID of PI + + module purge + module load wulver + module load foss/2025a GROMACS/2025.2-CUDA-12.8.0 + + INPUT_DIR=${PWD}/INPUT + OUTPUT_DIR=${PWD}/OUTPUT + + cp -r $INPUT_DIR/* $OUTPUT_DIR/ + cd $OUTPUT_DIR + + srun gmx_mpi mdrun -deffnm run -cpi -v -ntomp 1 -pin on -tunepme -dlb yes -nb gpu -noappend + ``` + + === "CPU" + + ```slurm + #!/bin/bash -l + # NOTE the -l (login) flag! + #SBATCH -J gmx2021 + #SBATCH -o test.%x.%j.out + #SBATCH -e test.%x.%j.err + #SBATCH --mail-type=ALL + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --time 72:00:00 # Max 3 days + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --account=$PI_ucid # Replace PI_ucid with the UCID of PI + + module purge + module load wulver + module load foss/2025a GROMACS/2025.2 + + INPUT_DIR=${PWD}/INPUT + OUTPUT_DIR=${PWD}/OUTPUT + + cp -r $INPUT_DIR/* $OUTPUT_DIR/ + cd $OUTPUT_DIR + + srun gmx_mpi mdrun -v -deffnm em -cpi -v -ntomp 1 -pin on -tunepme -dlb yes -noappend + ``` + + === "CPU with threads" + + ```slurm + #!/bin/bash -l + # NOTE the -l (login) flag! + #SBATCH -J gmx2021 + #SBATCH -o test.%x.%j.out + #SBATCH -e test.%x.%j.err + #SBATCH --mail-type=ALL + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --time 72:00:00 # Max 3 days + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=8 + #SBATCH --cpus-per-task=2 + #SBATCH --account=$PI_ucid # Replace PI_ucid with the UCID of PI + + module purge + module load wulver + module load foss/2025a GROMACS/2025.2 + + INPUT_DIR=${PWD}/INPUT + OUTPUT_DIR=${PWD}/OUTPUT + OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK + + cp -r $INPUT_DIR/* $OUTPUT_DIR/ + cd $OUTPUT_DIR + + srun gmx_mpi mdrun -v -deffnm em -cpi -v -ntomp $SLURM_CPUS_PER_TASK -pin on -tunepme -dlb yes -noappend + ``` + + +The tutorial in the above-mentioned job script can be found in `/apps/testjobs/gromacs` + + +## Related Applications + +* [LAMMPS](lammps.md) + +## User Contributed Information + +!!! info "Please help us improve this page" + + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). + + diff --git a/docs/Software/md/lammps.md b/docs/Software/Molecular_Dynamics/lammps.md similarity index 66% rename from docs/Software/md/lammps.md rename to docs/Software/Molecular_Dynamics/lammps.md index b344c07eb..581b2cc1f 100644 --- a/docs/Software/md/lammps.md +++ b/docs/Software/Molecular_Dynamics/lammps.md @@ -13,17 +13,7 @@ parallel particle simulator at the atomic, meso, or continuum scale. ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "LAMMPS"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "LAMMPS"') print(soft.to_markdown(index=False)) ``` @@ -35,11 +25,7 @@ parallel particle simulator at the atomic, meso, or continuum scale. ## Application Information, Documentation and Support -The official LAMMPS is available at [LAMMPS Online Manual](https://lammps.sandia.gov/doc/Manual.html). -LAMMPS has a large user base and a good user support. -Question related to using LAMMPS can be posted to the [LAMMPS User forum](https://matsci.org/c/lammps/40). -Archived [user mailing list](https://sourceforge.net/p/lammps/mailman/lammps-users/) are also useful to resolve -some of the common user issues. +The official LAMMPS is available at [LAMMPS Online Manual](https://lammps.sandia.gov/doc/Manual.html). LAMMPS has a large user base and a good user support. Question related to using LAMMPS can be posted to the [LAMMPS User forum](https://matsci.org/c/lammps/40). Archived [user mailing list](https://sourceforge.net/p/lammps/mailman/lammps-users/) are also useful to resolve some of the common user issues. !!! tip @@ -51,7 +37,7 @@ some of the common user issues. ??? example "Sample Batch Script to Run LAMMPS" - === "Wulver" + === "CPU" ```slurm #!/bin/bash @@ -61,7 +47,7 @@ some of the common user issues. #SBATCH --partition=general #SBATCH --nodes=1 #SBATCH --ntasks-per-node=128 - #SBATCH --mem-per-cpu=4000M # Maximum allowable mempry per CPU 4G + #SBATCH --mem-per-cpu=4000M # Maximum allowable memory per CPU 4G #SBATCH --qos=standard #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI #SBATCH --time=71:59:59 # D-HH:MM:SS @@ -73,32 +59,41 @@ some of the common user issues. ################################################ module purge module load wulver # Load slurm, easybuild - module load foss/2021b LAMMPS + module load foss/2024a LAMMPS - srun -n $SLURM_NTASKS lmp -in test.in + srun lmp -in test.in ``` - - === "Lochness" - + === "GPU" + ```slurm #!/bin/bash - #SBATCH -J test_lammps + #SBATCH -J gpu_lammps #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID - #SBATCH --partition=public - #SBATCH --nodes=2 - #SBATCH --ntasks-per-node=32 - #SBATCH --mem-per-cpu=10G # Adjust as necessary - #SBATCH --time=00:01:00 # D-HH:MM:SS - + #SBATCH --error=%x.%j.err # prints the error message + #SBATCH --partition=gpu + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=128 + #SBATCH --mem-per-cpu=4000M # Maximum allowable memory per CPU 4G + #SBATCH --qos=standard + #SBATCH --gres=gpu:2 + #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI + #SBATCH --time=71:59:59 # D-HH:MM:SS + ############################################### # # Purge and load modules needed for run # ################################################ module purge - module load foss/2021b LAMMPS + module load wulver # Load slurm, easybuild + module load foss/2024a LAMMPS/29Aug2024_update2-kokkos-CUDA-12.6.0 + + export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK + export OMP_PROC_BIND=spread + export OMP_PLACES=threads + - srun -n $SLURM_NTASKS lmp -in test.in + srun lmp -in in.lj -k on g 2 -sf kk -pk kokkos newton off neigh full comm device ``` Then submit the job script using the sbatch command, e.g., assuming the job script name is `test_lammps.slurm`: @@ -140,28 +135,6 @@ or from the [LAMMPS Github repository](https://github.com/lammps/lammps). make install ``` - === "Lochness" - - ```bash - module purge - module load foss - module load CMake - - git clone https://github.com/lammps/lammps.git - cd lammps - mkdir build - cd build - - cmake -DCMAKE_INSTALL_PREFIX=$PWD/../install_hsw -DCMAKE_CXX_COMPILER=mpicxx \ - -DCMAKE_BUILD_TYPE=Release -D BUILD_MPI=yes -DKokkos_ENABLE_OPENMP=ON \ - -DKokkos_ARCH_HSW=ON -DCMAKE_CXX_STANDARD=17 -D PKG_MANYBODY=ON \ - -D PKG_MOLECULE=ON -D PKG_KSPACE=ON -D PKG_REPLICA=ON -D PKG_ASPHERE=ON \ - -D PKG_RIGID=ON -D PKG_KOKKOS=ON -D DOWNLOAD_KOKKOS=ON \ - -D CMAKE_POSITION_INDEPENDENT_CODE=ON -D CMAKE_EXE_FLAGS="-dynamic" ../cmake - make -j16 - make install - ``` - ## Related Applications * [GROMACS](gromacs.md) @@ -170,7 +143,7 @@ or from the [LAMMPS Github repository](https://github.com/lammps/lammps). !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/md/plumed.md b/docs/Software/Molecular_Dynamics/plumed.md similarity index 56% rename from docs/Software/md/plumed.md rename to docs/Software/Molecular_Dynamics/plumed.md index 794169f56..5eea04a04 100644 --- a/docs/Software/md/plumed.md +++ b/docs/Software/Molecular_Dynamics/plumed.md @@ -9,29 +9,20 @@ title: PLUMED ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "PLUMED"') print(soft.to_markdown(index=False)) ``` -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "PLUMED"') - print(soft.to_markdown(index=False)) - ``` ## Related Applications -* +* [GROMACS](gromacs.md), [LAMMPS](lammps.md) ## User Contributed Information !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/cp2k.md b/docs/Software/chemistry/cp2k.md index e7ae4be5c..1816d8ce5 100644 --- a/docs/Software/chemistry/cp2k.md +++ b/docs/Software/chemistry/cp2k.md @@ -11,39 +11,64 @@ CP2K is widely used in the fields of materials science, chemistry, and physics f ## Availability -=== "Wulver" +```python exec="on" +import pandas as pd - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "CP2K"') - print(soft.to_markdown(index=False)) - ``` +df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') +soft = df.query('Software == "CP2K"') +print(soft.to_markdown(index=False)) +``` -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "CP2K"') - print(soft.to_markdown(index=False)) - ``` ## Application Information, Documentation The documentation of CP2K is available at [CP2K Documentation](https://www.cp2k.org/docs). For any issues CP2K simulation, users can contact at [CP2K Forum](https://www.cp2k.org/howto:forum). ## Using CP2K CP2K MPI/OpenMP-hybrid Execution (PSMP), CP2K with Population Analysis capabilities- CP2K-popt + +??? example "Sample Batch Script to Run CP2K : cp2k.submit.sh" + + ```slurm + #!/bin/bash -l + #SBATCH -J CP2K + #SBATCH -o sn1-xtb_input.out + #SBATCH -e sn1-xtb_input.err + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=16 + #SBATCH --mem-per-cpu=4G + #SBATCH --qos=standard + #SBATCH --partition=general + #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI + #SBATCH -t 72:00:00 + + #module load command + + module purge > /dev/null 2>&1 + module load wulver + module load foss/2025a CP2K + + #Run the program + + inputFile=sn1-xtb_input.inp + outputFile=sn1-xtb_input.out + mpirun -np $SLURM_NTASKS cp2k.popt -i $inputFile -o $outputFile + ``` + +The sample input file `sn1-xtb_input.inp` can be found in `/apps/testjobs/CP2K` + +!!! important + + Please don't run anything on `/apps/testjobs/CP2K` as users have only read-only permission. You need to copy the submit script and input file from `/apps/testjobs/CP2K` to `$HOME` or `/project`. + ## Related Applications -* +* [Gaussian](gaussian.md) +* [ORCA](orca.md) ## User Contributed Information !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/gaussian.md b/docs/Software/chemistry/gaussian.md index 857fdb568..58a3fe5d3 100644 --- a/docs/Software/chemistry/gaussian.md +++ b/docs/Software/chemistry/gaussian.md @@ -10,29 +10,62 @@ title: Gaussian ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "Gaussian"') print(soft.to_markdown(index=False)) ``` -=== "Lochness" +!!! important - ```python exec="on" - import pandas as pd + Due to licensing restrictions, Gaussian is not automatically accessible to all HPC users. Students are required to contact [hpc@njit.edu](mailto:hpc@njit.edu) to request access to Gaussian. + +## Application Information, Documentation +The documentation of Gaussian is available at [Gaussian Documentation](https://gaussian.com/man/). For any issues Gaussian simulation, users can contact at [Gaussian Support](https://gaussian.com/techsupport/). + +## Using Gaussian +??? example "Sample Batch Script to Run Gaussian : g16.submit.sh" - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "Gaussian" | Software == "gaussian"') - print(soft.to_markdown(index=False)) - ``` + === "Wulver" + + ```slurm + #!/bin/bash -l + #SBATCH -J g16 + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=4 + #SBATCH --mem-per-cpu=3G + #SBATCH --time=10:00:00 + #SBATCH --partition=general + #SBATCH --account=PI_UCID # Replace PI_UCID with the UCID of PI + #SBATCH --output=%x.%j.out # %x.%j expands to slurm JobName.JobID + #SBATCH --error=%x.%j.err + #SBATCH --qos=standard + + #module load command + + module purge > /dev/null 2>&1 + module load wulver + module load Gaussian + + #Run the program + + g16 test_g98.com + ``` + +The sample input file `test_g98.com` can be found in `/apps/testjobs/Gaussian` + +!!! important + + Please don't run anything on `/apps/testjobs/Gaussian` as users have only read-only permission. You need to copy the submit script and input file from `/apps/testjobs/Gaussian` to `$HOME` or `/project`. ## Related Applications -* +* [CP2K](cp2k.md) +* [ORCA](orca.md) ## User Contributed Information !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/orca.md b/docs/Software/chemistry/orca.md index 44a8f1c68..dde377b9e 100644 --- a/docs/Software/chemistry/orca.md +++ b/docs/Software/chemistry/orca.md @@ -9,30 +9,58 @@ title: ORCA ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "ORCA"') print(soft.to_markdown(index=False)) ``` -=== "Lochness" +## Application Information, Documentation +The documentation of Gaussian is available at [ORCA Documentation](https://www.orcasoftware.de/tutorials_orca/). For any issues Gaussian simulation, users can contact at [ORCA Forum](https://orcaforum.kofo.mpg.de/app.php/portal). - ```python exec="on" - import pandas as pd +## Using Gaussian +??? example "Sample Batch Script to Run ORCA : orca.submit.sh" - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "ORCA"') - print(soft.to_markdown(index=False)) - ``` + === "Wulver" + + ```slurm + #!/bin/bash -l + #SBATCH --job-name=job_orca + #SBATCH --output=%x.%j.out + #SBATCH --error=%x.%j.err + #SBATCH --partition=general + #SBATCH --qos=standard + #SBATCH --nodes=1 + #SBATCH --account=PI_UCID # Replace PI_UCID with the UCID of PI + #SBATCH --ntasks-per-node=8 + #SBATCH --time=59:00 # D-HH: + + #module load command + + module purge > /dev/null 2>&1 + module load wulver + module load foss/2024a ORCA + + #Run the program + + srun orca test.inp > geom.out + ``` + +The sample input file `test.inp` can be found in `/apps/testjobs/ORCA` + +!!! important + + Please don't run anything on `/apps/testjobs/ORCA` as users have only read-only permission. You need to copy the submit script and input file from `/apps/testjobs/ORCA` to `$HOME` or `/project`. ## Related Applications -* +* [Gaussian](gaussian.md) +* [CP2K](cp2k.md) ## User Contributed Information !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/qe.md b/docs/Software/chemistry/qe.md index d36c80394..7b9a43a8e 100644 --- a/docs/Software/chemistry/qe.md +++ b/docs/Software/chemistry/qe.md @@ -9,17 +9,7 @@ title: Quantum Espresso ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "QuantumESPRESSO"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "QuantumESPRESSO"') print(soft.to_markdown(index=False)) ``` @@ -32,7 +22,7 @@ title: Quantum Espresso !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/siesta.md b/docs/Software/chemistry/siesta.md index 1ed660d97..20a58661e 100644 --- a/docs/Software/chemistry/siesta.md +++ b/docs/Software/chemistry/siesta.md @@ -14,17 +14,7 @@ SIESTA is actively developed and maintained by a team of researchers at the Univ ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "Siesta"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "Siesta"') print(soft.to_markdown(index=False)) ``` @@ -37,7 +27,7 @@ SIESTA is actively developed and maintained by a team of researchers at the Univ !!! info "Please help us improve this page" - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). diff --git a/docs/Software/chemistry/vasp.md b/docs/Software/chemistry/vasp.md new file mode 100644 index 000000000..47cbf8a93 --- /dev/null +++ b/docs/Software/chemistry/vasp.md @@ -0,0 +1,61 @@ +--- +title: VASP +--- +VASP (Vienna Ab initio Simulation Package) is a commercial software package for performing first-principles quantum mechanical calculations in materials science, chemistry, and physics. It is based primarily on density functional theory (DFT), with extensions for time-dependent DFT and many-body perturbation theory (e.g., GW and RPA). + +VASP is particularly known for its efficient and accurate implementation of the projector augmented-wave (PAW) method, enabling precise electronic structure calculations. It supports geometry optimizations, molecular dynamics, phonon calculations, and band structure analysis, making it a powerful tool for studying molecules, solids, surfaces, and interfaces. + +The code is optimized for massively parallel supercomputers and includes advanced iterative diagonalization and charge-density mixing algorithms for large-scale simulations. Although VASP itself does not include a GUI, it integrates seamlessly with a wide range of external tools for input generation and visualization of results. + +## Availability + +Since VASP is **licensed software**, access is restricted to the cluster. If your research group has a valid VASP license and wants to use it, please ask your advisor/PI to email [hpc@njit.edu](mailto:hpc@njit.edu) with the following details + +1. UCIDs of the students who need access to VASP +2. Licensed VASP version (e.g., 6.x or 5.x) +3. Proof of license (license confirmation/contract) + + +## Application Information, Documentation +The documentation of CP2K is available at [VASP Documentation](https://vasp.at/wiki/The_VASP_Manual). For any issues in VASP simulation, users can contact at [VASP Forum](https://www.vasp.at/forum/). + +## Using VASP + +CP2K MPI/OpenMP-hybrid Execution (PSMP), CP2K with Population Analysis capabilities- CP2K-popt + +??? example "Sample Batch Script to Run CP2K : cp2k.submit.sh" + + ```slurm + #!/bin/bash -l + #SBATCH -J VASP + #SBATCH -o vasp.out + #SBATCH -e vasp.err + #SBATCH --nodes=1 + #SBATCH --ntasks-per-node=16 + #SBATCH --mem-per-cpu=4G + #SBATCH --qos=standard + #SBATCH --partition=general + #SBATCH --account=PI_ucid # Replace PI_ucid which the NJIT UCID of PI + #SBATCH -t 72:00:00 + + #module load command + + module purge > /dev/null 2>&1 + module load wulver + module load intel/2025a HDF5 VASP + + srun --mpi=pmix vasp_std > vasp.log + ``` + +## Related Applications + +* [Gaussian](gaussian.md) +* [ORCA](orca.md) + +## User Contributed Information + +!!! info "Please help us improve this page" + + Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/issues). + + diff --git a/docs/Software/index.md b/docs/Software/index.md index 15cdd5caf..81dcb73fe 100644 --- a/docs/Software/index.md +++ b/docs/Software/index.md @@ -1,63 +1,75 @@ # Software Environment -All software and numerical libraries available at the cluster can be found at `/opt/site/easybuild/software/` if you are using Lochness. In case of Wulver the applications are installed at `/apps/easybuild/software/`. We use [EasyBuild](https://docs.easybuild.io/en/latest) to install, build and manage different version of packages. -!!! note +All software and numerical libraries available at the cluster can be found at `/apps/easybuild/software/`. We use [EasyBuild](https://docs.easybuild.io/en/latest) to install, build and manage different version of packages. - If you are using Lochness, add the following in `.modules` file located in `$HOME` directory - ``` - module use /opt/site/easybuild/modules/all/MPI - module use /opt/site/easybuild/modules/all/Core - module use /opt/site/easybuild/modules/all/Compiler - ``` -If you could not find software or libraries on HPC cluster, please contact us at [hpc@njit.edu](mailto:hpc@njit.edu) to get it installed. -The installed software or packages can be accessed as [Lmod](https://lmod.readthedocs.io/en/latest) modules. -## Modules +If you could not find software or libraries on HPC cluster, please submit a request for [HPC Software Installation](https://njit.service-now.com/sp?id=sc_cat_item&sys_id=0746c1f31b6691d04c82cddf034bcbe2&sysparm_category=405f99b41b5b1d507241400abc4bcb6b) by visiting the [Service Catalog](https://njit.service-now.com/sp?id=sc_category). The list of installed software or packages on HPC cluster can be found in [Software List](#software-list). -### Access to All Available Modules -The list of software and libraries installed on Lochness can be accessed by using the following command +!!! warning + + When installing software on Wulver, please do not use `sudo` under any circumstances. The use of `sudo` is restricted to system administrators and is not permitted in user environments for security and stability reasons. Users should install software in their own directories using tools like [conda](conda.md), pip, or by compiling from source in their home or project space. You may come across the use of `sudo` in some software installation instructions, but please note that these are intended for personal computers only. If you require assistance with installation of a package, please contact the [HPC support](mailto:hpc@njit.edu) team or build the package in an [apptainer](apptainer.md) container. + +## Modules +We use Environment Modules to manage the user environment in HPC, which help users to easily load and unload software packages, switch between different versions of software, and manage complex software dependencies. [Lmod](https://lmod.readthedocs.io/en/latest) is an extension of the Environment Modules system, implemented in Lua. It enhances the functionality of traditional Environment Modules by introducing features such as hierarchical module naming, module caching, and improved flexibility in managing environment variables. -``` -module av -``` ### Search for Specific Package -You can check specific packages and list of their versions using `module av`. For example, the list of different versions of Python installed on cluster cna be checked by using, -```console -module av Python -``` -The above command gives the following output +You can check specific packages and list of their versions using `module spider`. For example, the list of Python installed on cluster can be checked by using + ```console -------------------------------------------------------------------------------------------------------------------- /opt/site/easybuild/modules/all/Compiler ------------------------------------------------------------------------------------------------------------------- - GCCcore/10.2.0/Python/2.7.18 GCCcore/11.2.0/IPython/7.26.0 GCCcore/8.3.0/Meson/0.51.2-Python-3.7.4 GCCcore/9.3.0/Flask/1.1.2-Python-3.8.2 GCCcore/9.3.0/archspec/0.1.0-Python-3.8.2 - GCCcore/10.2.0/Python/3.8.6 (D) GCCcore/11.2.0/Python/2.7.18-bare GCCcore/8.3.0/Meson/0.59.1-Python-3.7.4 (D) GCCcore/9.3.0/GObject-Introspection/1.64.0-Python-3.8.2 GCCcore/9.3.0/pkgconfig/1.5.1-Python-3.8.2 - GCCcore/10.3.0/Python/2.7.18-bare GCCcore/11.2.0/Python/3.9.6-bare GCCcore/8.3.0/Python/2.7.16 GCCcore/9.3.0/Meson/0.55.1-Python-3.8.2 GCCcore/9.3.0/pybind11/2.4.3-Python-3.8.2 - GCCcore/10.3.0/Python/3.9.5-bare GCCcore/11.2.0/Python/3.9.6 (D) GCCcore/8.3.0/Python/3.7.4 (D) GCCcore/9.3.0/Python/2.7.18 - GCCcore/10.3.0/Python/3.9.5 (D) GCCcore/11.2.0/protobuf-python/3.17.3 GCCcore/8.3.0/pkgconfig/1.5.1-Python-3.7.4 GCCcore/9.3.0/Python/3.8.2 (D) -``` -!!! note +module spider Python + +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- + Description: + Python is a programming language that lets you work more quickly and integrate your systems more effectively. + + Versions: + Python/2.7.18-bare + Python/3.9.6-bare + Python/3.9.6 + Python/3.10.4-bare + Python/3.10.4 + Python/3.10.8-bare + Python/3.10.8 + Python/3.11.5 + Other possible modules matches: + Biopython Boost.Python Python-bundle-PyPI meson-python python3 python39 + +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- + To find other possible module matches execute: - The above display message is for Lochness only. In Wulver, most of the appications are built based on the toolchain. For more details see [Toolchain](compilers.md#toolchains). Therefore to see available software, you need to load the toolchain first. + $ module -r spider '.*Python.*' +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- + For detailed information about a specific "Python" package (including how to load the modules) use the module's full name. + Note that names that have a trailing (E) are extensions provided by other modules. + For example: + + $ module spider Python/3.11.5 +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- + + +``` +This will show the different versions of Python available on Wulver. + +To see how to load the specific version of software (for example `Python/3.9.6`), the following command needs to be used. -To see how to load the modules (for example `Python/3.9.6`) the following command needs to used. ```console module spider Python/3.9.6 ``` -This will show the which prerequisite modules need to loaded prior to loading `Python` +This will show the which prerequisite modules need to be loaded prior to loading `Python/3.9.6` ```console ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Python: Python/3.9.6 ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- +--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Description: Python is a programming language that lets you work more quickly and integrate your systems more effectively. You will need to load all module(s) on any one of the lines below before the "Python/3.9.6" module is available to load. - Core/GCCcore/11.2.0 - GCCcore/11.2.0 + easybuild GCCcore/11.2.0 + slurm/wulver GCCcore/11.2.0 Help: - Description =========== Python is a programming language that lets you work more quickly and integrate your systems @@ -71,17 +83,18 @@ This will show the which prerequisite modules need to loaded prior to loading `P If you are unsure about the version, you can also use `module spider Python` to see the different versions of Python and prerequisite modules to be loaded. ### Load Modules -To use specific package you need to use `module load` command which modified the environment to load the software package(s). +To use specific package, you need to use `module load` command which modified the environment to load the software package(s). !!! Note * The `module load` command will load dependencies automatically as needed, however you may still need to load prerequisite modules to load specific software package(s). For that you need to use `module spider` command as described above. * For running jobs via batch script, you need to add module load command(s) to your submission script. -For example, to load `Python` version `3.9.6` as shown in the above example, you need to load `GCCcore/.11.2.0` module first before loading the Python module is available to load. To use `Python 3.9.6`, use the following command + +For example, to load `Python` version `3.9.6` as shown in the above example, you need to load `GCCcore/11.2.0` module first before loading the Python module is available to load. To use `Python 3.9.6`, use the following command ```console module load GCCcore/11.2.0 Python ``` -You can verify whether Python is loaded using, +You can verify whether Python is loaded using ```console module li @@ -89,11 +102,8 @@ module li and this will result is the following output ```console Currently Loaded Modules: - 1) GCCcore/11.2.0 (H) 3) binutils/2.37 (H) 5) ncurses/6.2 (H) 7) Tcl/8.6.11 9) XZ/.5.2.5 (H) 11) libffi/3.4.2 (H) 13) Python/3.9.6 - 2) zlib/1.2.11 (H) 4) bzip2/1.0.8 (H) 6) libreadline/8.1 (H) 8) SQLite/3.36 10) GMP/.6.2.1 (H) 12) OpenSSL/1.1 (H) + 1) easybuild 2) wulver 3) slurm/wulver 4) null 5) GCCcore/11.2.0 6) zlib/1.2.11 7) binutils/2.37 8) bzip2/1.0.8 9) ncurses/6.2 10) libreadline/8.1 11) Tcl/8.6.11 12) SQLite/3.36 13) XZ/5.2.5 14) GMP/6.2.1 15) libffi/3.4.2 16) OpenSSL/1.1 17) Python/3.9.6 - Where: - H: Hidden Module ``` ### Module unload @@ -156,19 +166,21 @@ man module The following applications are installed on Wulver. -=== "Wulver" +=== "Wulver RHEL9" ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') + df = df.sort_values(by=df.columns[0], key=lambda col: col.str.lower()) # Sorts by the first column alphabetically print(df.to_markdown(index=False)) ``` - -=== "Lochness" +=== "Wulver RHEL8" ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel8.csv') + df = df.sort_values(by=df.columns[0], key=lambda col: col.str.lower()) # Sorts by the first column alphabetically print(df.to_markdown(index=False)) ``` + diff --git a/docs/Software/math/MATLAB/img/GenericProfile5.png b/docs/Software/math/MATLAB/img/GenericProfile5.png deleted file mode 100644 index 270ca9995..000000000 Binary files a/docs/Software/math/MATLAB/img/GenericProfile5.png and /dev/null differ diff --git a/docs/Software/math/MATLAB/img/GenericProfile7.png b/docs/Software/math/MATLAB/img/GenericProfile7.png deleted file mode 100644 index 32b600d05..000000000 Binary files a/docs/Software/math/MATLAB/img/GenericProfile7.png and /dev/null differ diff --git a/docs/Software/math/MATLAB/img/GenericProfile9.png b/docs/Software/math/MATLAB/img/GenericProfile9.png deleted file mode 100644 index 94b00645a..000000000 Binary files a/docs/Software/math/MATLAB/img/GenericProfile9.png and /dev/null differ diff --git a/docs/Software/math/MATLAB/matlab_local.md b/docs/Software/math/MATLAB/matlab_local.md deleted file mode 100644 index c48c7a0da..000000000 --- a/docs/Software/math/MATLAB/matlab_local.md +++ /dev/null @@ -1,270 +0,0 @@ -# Use MATLAB on NJIT HPC - -!!! warning - - Please note that the following instructions are applicatible for Lochness only. We will soon update the instructions for Wulver. - -## Installation steps of MATLAB on local machine -* Go to [Mathworks Download](https://www.mathworks.com/downloads/) and register with your NJIT email address. -* Select the **R2022a** version to download. -* User needs to select the correct installer based on the OS (Mac or Windows). -* Run the installer. - - { width=50% height=50%} - { width=50% height=50%} - -* Make sure to check **Parallel Computing Toolbox** option. - - {width=50% height=50%} - -* Continue by selecting **Next** and MATLAB will be installed on your computer. - -## Setup Slurm profile to run MATLAB on Lochness -Following this procedure a user will be able to submit jobs to lochness or stheno from Matlab running locally on the user's computer. - -### Installing the Add-On - -From the Matlab window, click on "Add-ons" and select "Get Add-Ons." - - { width=70% height=70%} - -In the search box enter "slurm" and click on the magnifying glass icon. Select "Parallel Computing Toolbox plugin for MATLAB Parallel Server with Slurm". Alternatively, this Add-On can be downloaded directly from the [Mathworks](https://www.mathworks.com/matlabcentral/fileexchange/52807-parallel-computing-toolbox-plugin-for-matlab-parallel-server-with-slurm) site. - - { width=70% height 70%} - -Click on "Install." - -{ width=70% height 70%} - -### Creating a Profile for Lochness or Stheno - -The following steps will create a profile for lochness (or stheno). Click Next to begin. - -{ width=50% height 50%} - -In the "Operating System" screen `Unix` is already selected. Click Next to continue. - -{ width=50% height 50%} - -This "Submission Mode" screen determines whether or not to use a `shared` or `nonshared` submission mode. Since Matlab installed on your personal computer or laptop does not use a shared job location storage, select "No" where indicated and click Next to continue. - -{ width=50% height 50%} - -Click Next to continue. - -{ width=50% height 50%} - -In the "Connection Details" screen, enter the cluster host, either "lochness.njit.edu" or "stheno.njit.edu." Enter your UCID for the username. Select "No" for the "Do you want to use an identity file to log in to the cluster" option and click next to continue. - -{ width=50% height 50%} - -In the "Cluster Details" screen enter the full path to the directory on lochness to store the Matlab job files. In the case the directory is $HOME/MDCS. MDCS stands for Matlab Distributed Computing Server. It is not necessary to name this directory MDCS. This directory can be named anything you wish. To determine the value of $HOME, logon to lochness. For details on how to Logon to Lochness from local computer please see this [link](https://hackmd.io/@absrocks/BJRlQtBVi). Once connected to Lochness run the following: - -``` - login-1-45 ~ >: echo $HOME - /home/g/guest24 -``` - -Make sure to check the box Use unique subfolders . Click Next to continue. - -{ width=50% height 50%} - -In the "Workers" screen enter `512` for the number of workers and `/opt/site/easybuild/software/MATLAB/2022a` for `MATLAB installation folders for workers`. Click Next to continue. - -{ width=50% height 50%} - -In the "License" screen make sure to select "Network license manager" and click Next to continue. - -{ width=50% height 50%} - -In the "Profile Details" screen enter either "Lochness" or "Stheno" depending on which cluster you are making a profile for. The "Cluster description" is optional and may be left blank. Click Next to continue. - -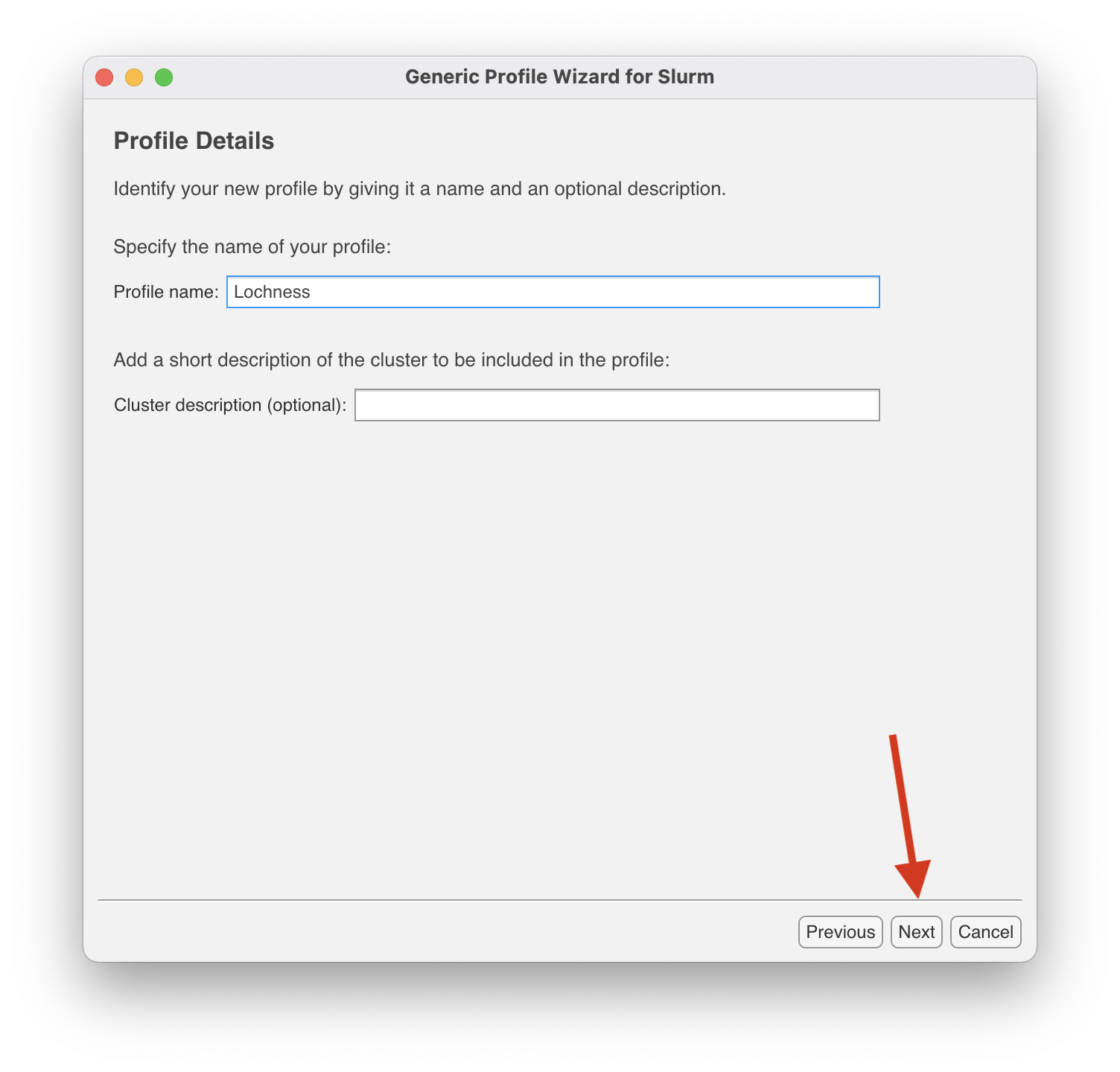{ width=50% height 50%} - -In the "Summary" screen make sure everything is correct and click "Create." - -{ width=50% height 50%} - -In the "Profile Created Successfully" screen, check the "Set the new profile as default" box and click on "Finish." - -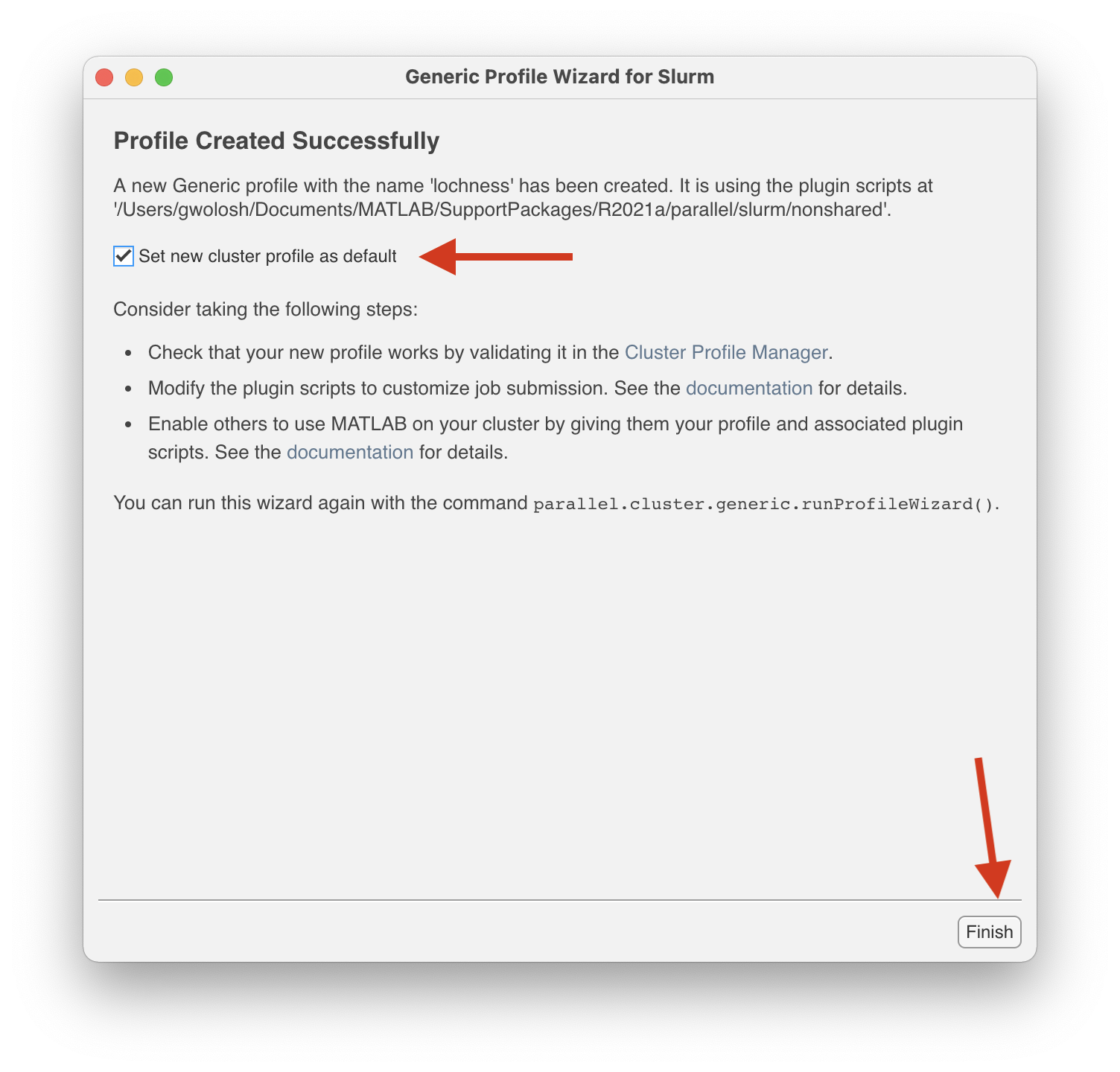{ width=50% height 50%} - -## Submitting a Serial Job -This section will demonstrate how to create a cluster object and submit a simple job to the cluster. The job will run the 'hostname' command on the node assigned to the job. The output will indicate clearly that the job ran on the cluster and not on the local computer. - -The hostname.m file used in this demonstration can be downloaded [here](https://www.mathworks.com/matlabcentral/fileexchange/24096-hostname-m). - -``` - >> c=parcluster -``` - 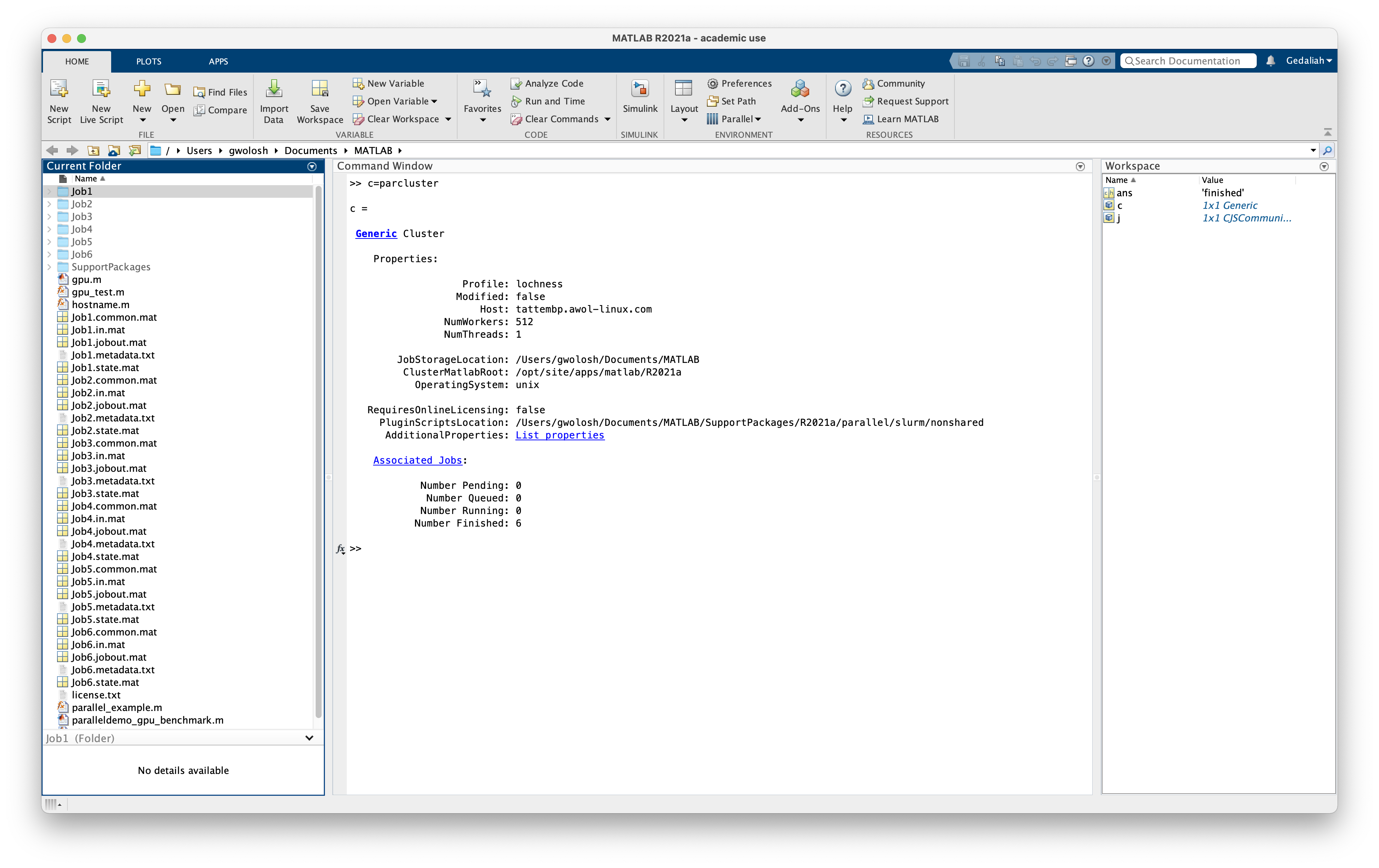{ width=70% height 70%} - -Certain arguments need to be passed to SLURM in order for the job to run properly. Here we will set values for partion, mem-per-cpu and time. In the Matlab window enter: -``` - >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=public --mem-per-cpu=10G --time=2-00:00:00'] -``` -To make this persistent between Matlab sessions these arguments need to be saved to the profile. In the Matlab window enter: -``` - >> c.saveProfile -``` - { width=70% height 70%} - -We will now submit the hostname.m function to the cluster. In the Matlab window enter the following: -``` ->> j=c.batch(@hostname, 1, {}, 'AutoAddClientPath', false); -``` -@: Submitting a function.\ -1: The number of output arguments from the evaluated function.\ -{}: Cell array of input arguments to the function. In this case empty.\ -'AutoAddClientPath', false: The client path is not available on the cluster. - - -When the job is submitted, you will be prompted for your password. - -For more information see the Mathworks page: [batch](https://www.mathworks.com/help/parallel-computing/batch.html) - -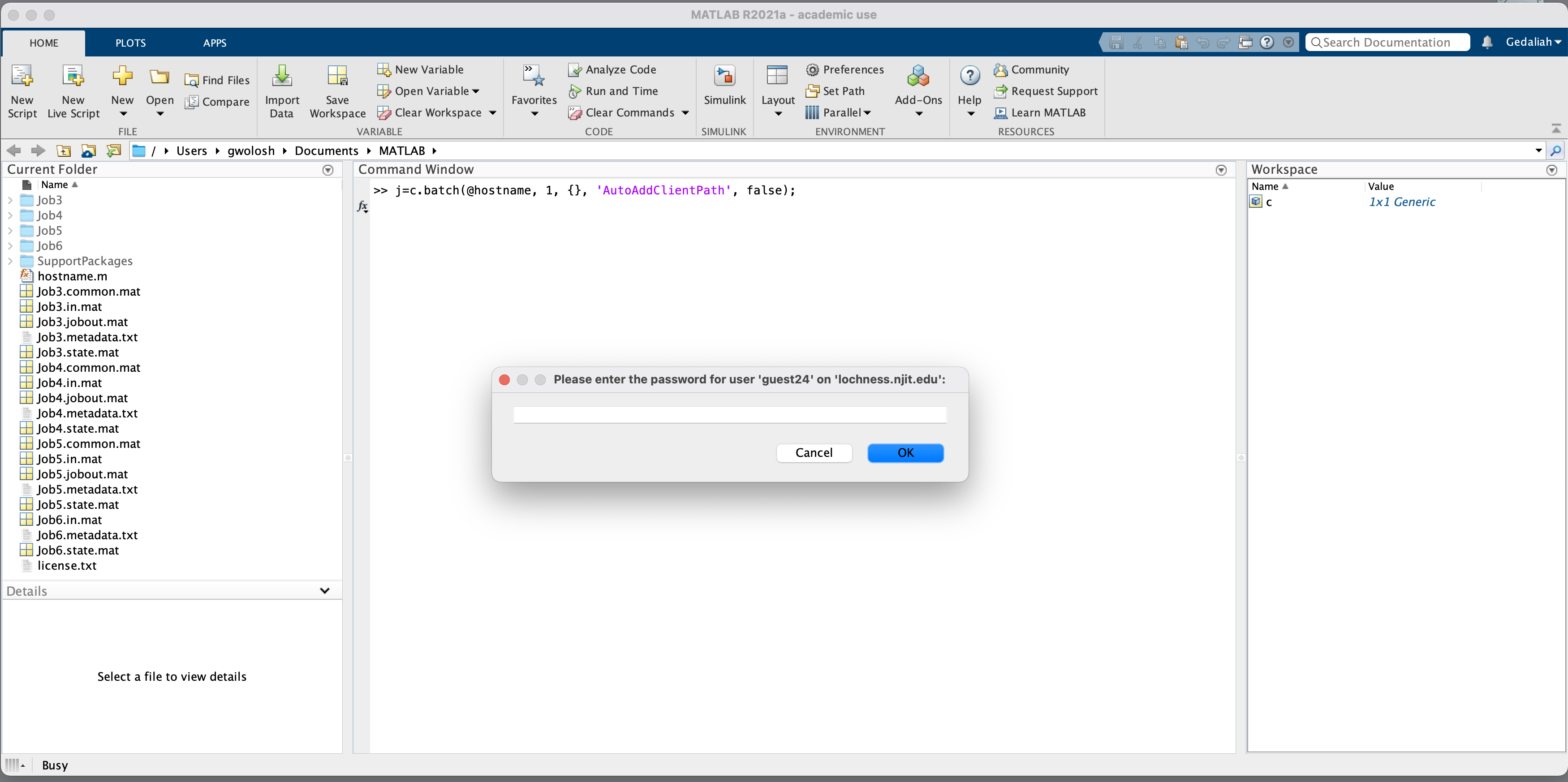{ width=70% height 70%} - -To wait for the job to finish, enter the following in the Matlab window: -``` - >>j.wait -``` -Finally, to get the results: -``` - >>fetchOutputs(j) -``` -As can be seen, this job ran on node720 - -{ align=left, width=70% height 70%} - -### Submitting a Parallel Function -The "Job Monitor" is a convenient way to monitor jobs submitted to the cluster. In the Matlab window select "Parallel" and then "Monitor Jobs." - -For more information see the Mathworks page: [Job Monitor](https://www.mathworks.com/help/parallel-computing/job-monitor.html) - -{ width=70% height 70%} - -Here we will submit a simple function using a "parfor" loop. The code for this example is as follows: -``` -function t = parallel_example - -t0 = tic; -parfor idx = 1:16 - A(idx) = idx; - pause (2) -end - -t=toc(t0); -``` -To submit this job: -``` - >> j=c.batch(@parallel_example, 1, {}, 'AutoAddClientPath', false, 'Pool', 7) -``` -Since this is a parallel job a 'Pool' must be started. The actual number of tasks started will be one more than requested in the pool. I this case, the batch command calls for a pool of seven. Eight tasks will be started on the cluster. - -Also see that the state of the job in the "Job Monitor" is "running." -{ width=70% height 70%} - -The job takes a few minutes to run and the state of the job changes to "finished." -{ width=70% height 70%} - -Once again to get the results enter: -``` - >> fetchOutputs(j) -``` -As can be seen the parfor loop was completed in 6.7591 seconds. -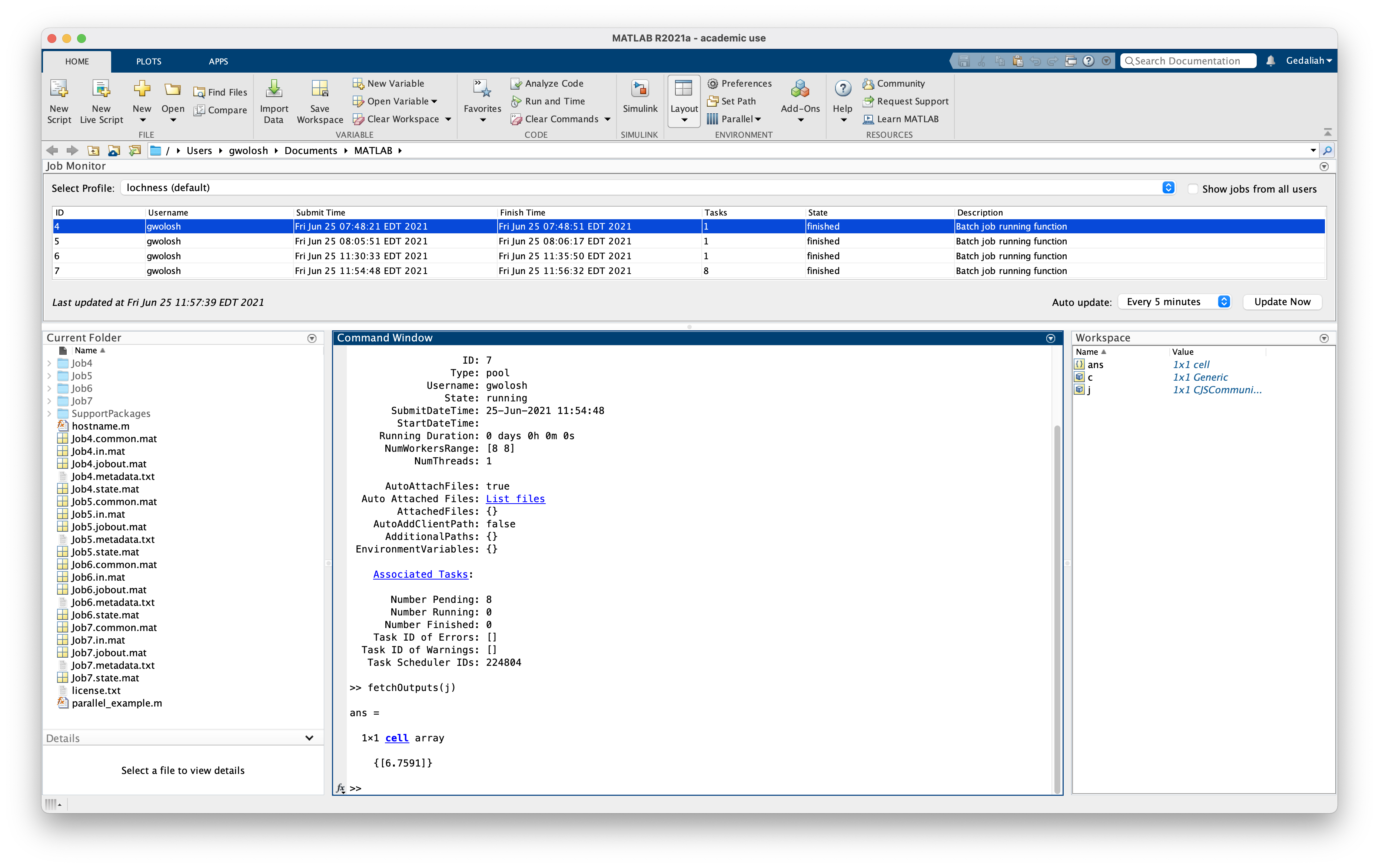{ width=70% height 70%} - -## Submitting a Script Requiring a GPU -In this section we will submit a matlab script using a GPU. The results will be written to the job diary. The code for this example is as follows: -``` -% MATLAB script that defines a random matrix and does FFT -% -% The first FFT is without a GPU -% The second is with the GPU -% -% MATLAB knows to use the GPU the second time because it -% is passed a type gpuArray as an argument to FFT -% We do the FFT a bunch of times to make using the GPU worth it, -% or else it spends more time offloading to the GPU -% than performning the calculation -% -% This example is meant to provide a general understanding -% of MATLAB GPU usage -% Meaningful performance measurements depend on many factors -% beyond the scope of this example -% Downloaded from https://projects.ncsu.edu/hpc/Software/examples/matlab/gpu/gpu_m - -% Define a matrix -A1 = rand(3000,3000); - -% Just use the compute node, no GPU -tic; -% Do 1000 FFT's -for i = 1:1000 - B2 = fft(A1); -end -time1 = toc; -fprintf('%s\n',"Time to run FFT on the node:") -disp(time1); - -% Use GPU -tic; -A2 = gpuArray(A1); -% Do 1000 FFT's -for i = 1:1000 - % MALAB knows to use GPU FFT because A2 is defined by gpuArray - B2 = fft(A2); -end -time2 = toc; -fprintf('%s\n',"Time to run FFT on the GPU:") -disp(time2); - -% Will be greater than 1 if GPU is faster -speedup = time1/time2 -``` -We will need to change the partition to datasci and request a gpu. In the Matlab window enter: -``` - >> c.AdditionalProperties.AdditionalSubmitArgs=['--partition=datasci --gres=gpu:1 --mem-per-cpu=10G --time=2-00:00:00'] -``` - - -Submit the job as before. Since a script is submitted as opposed to a function, only the name of the script is included in the batch command. Do not include the '@' symbol. In a script there are no inputs or ouptuts. -``` - >> j=c.batch('gpu', 'AutoAddClientPath', false) -``` - 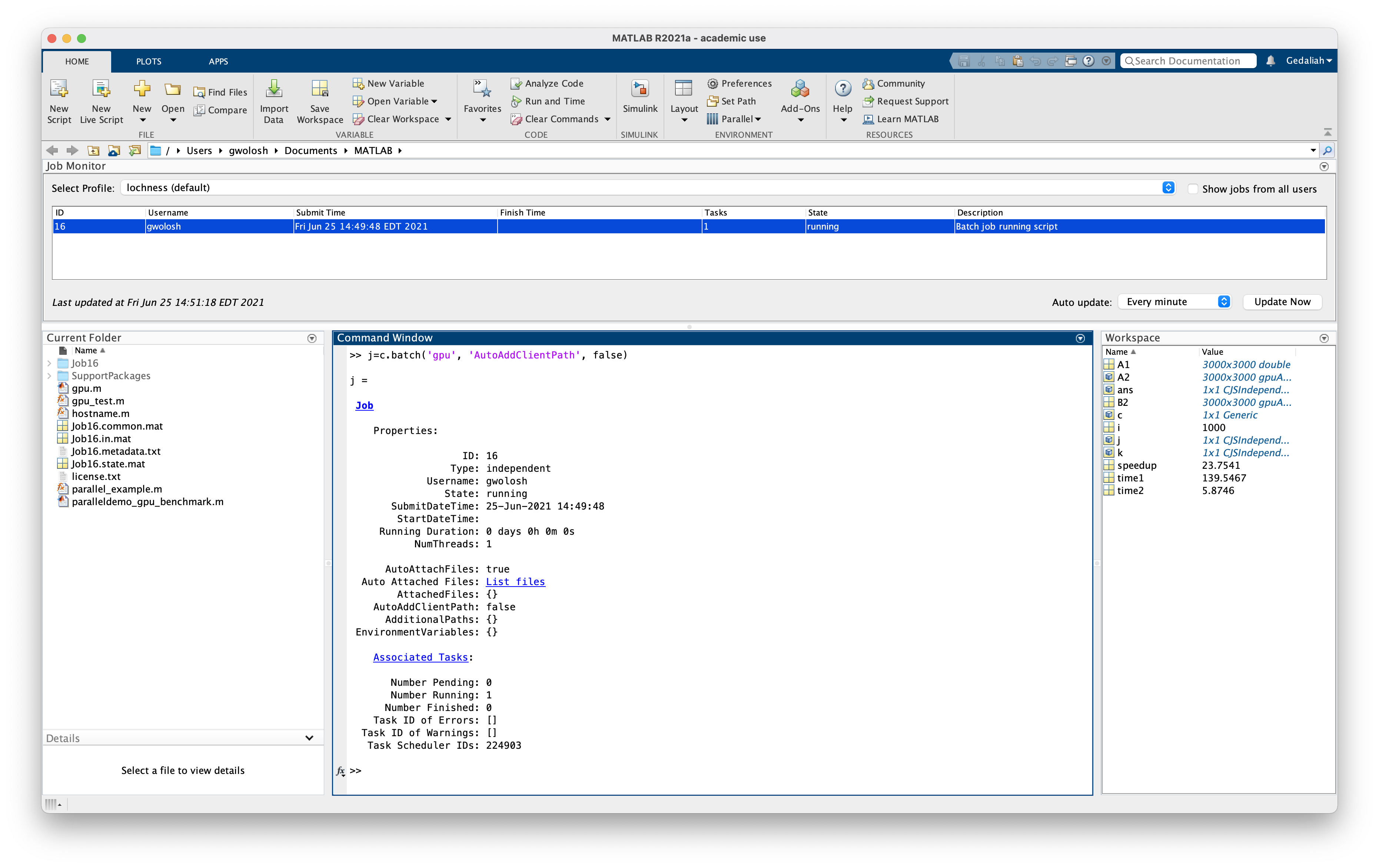{ width=70% height 70%} - -To get the result: -``` - >> j.diary -``` -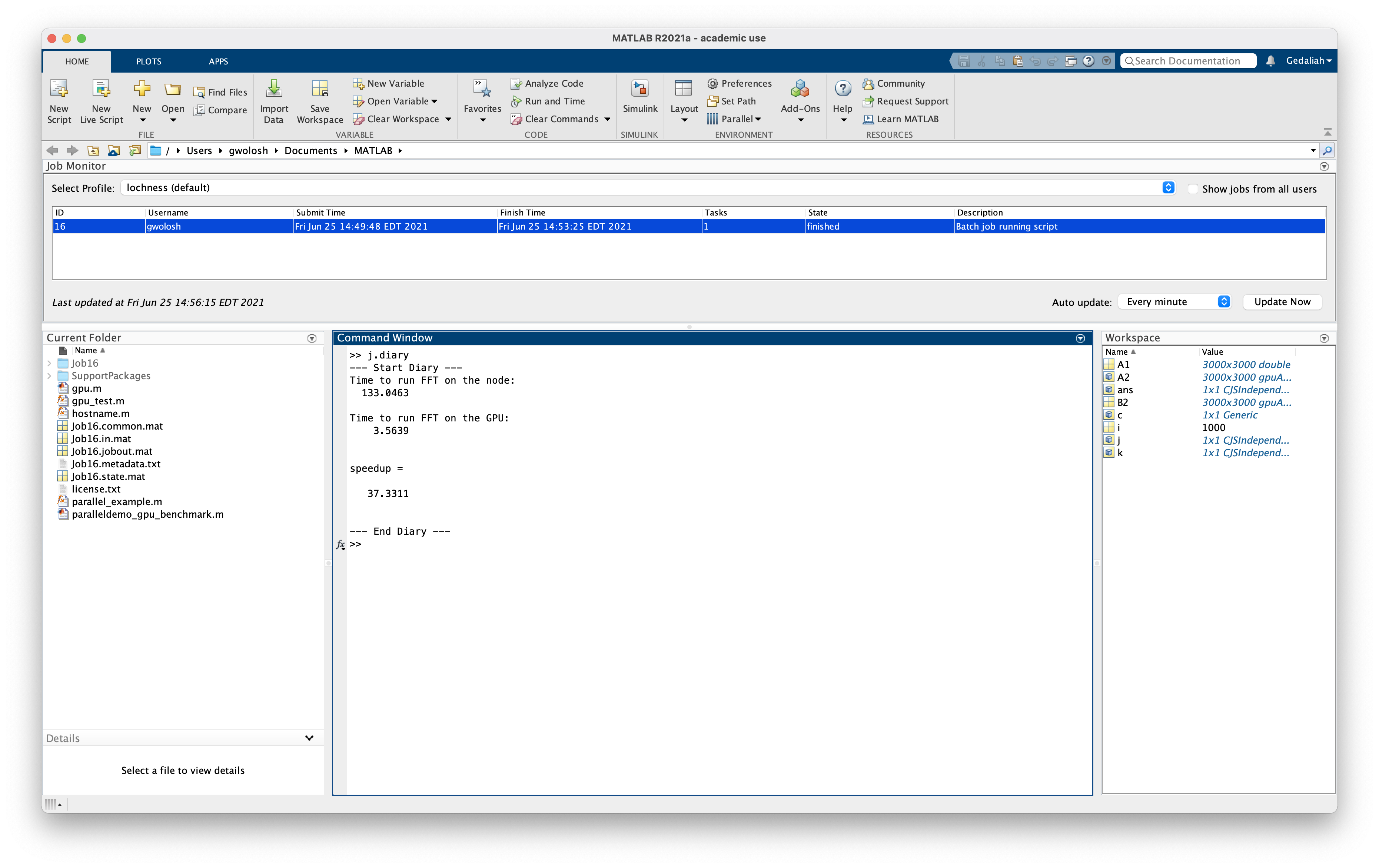{ width=70% height 70%} - -## Load and Plot Results from A Job -In this section we will run a job on the cluster and then load and plot the results in the local Matlab workspace. The code for this example is as follows: -``` -n=100; -disp("n = " + n); -A = gallery('poisson',n-2); -b = convn(([1,zeros(1,n-2),1]'|[1,zeros(1,n-1)]), 0.5*ones(3,3),'valid')'; -x = reshape(A\b(:),n-2,n-2)';% -``` -As before submit the job: -``` - >> j=c.batch('plot_demo', 'AutoAddClientPath', false); -``` -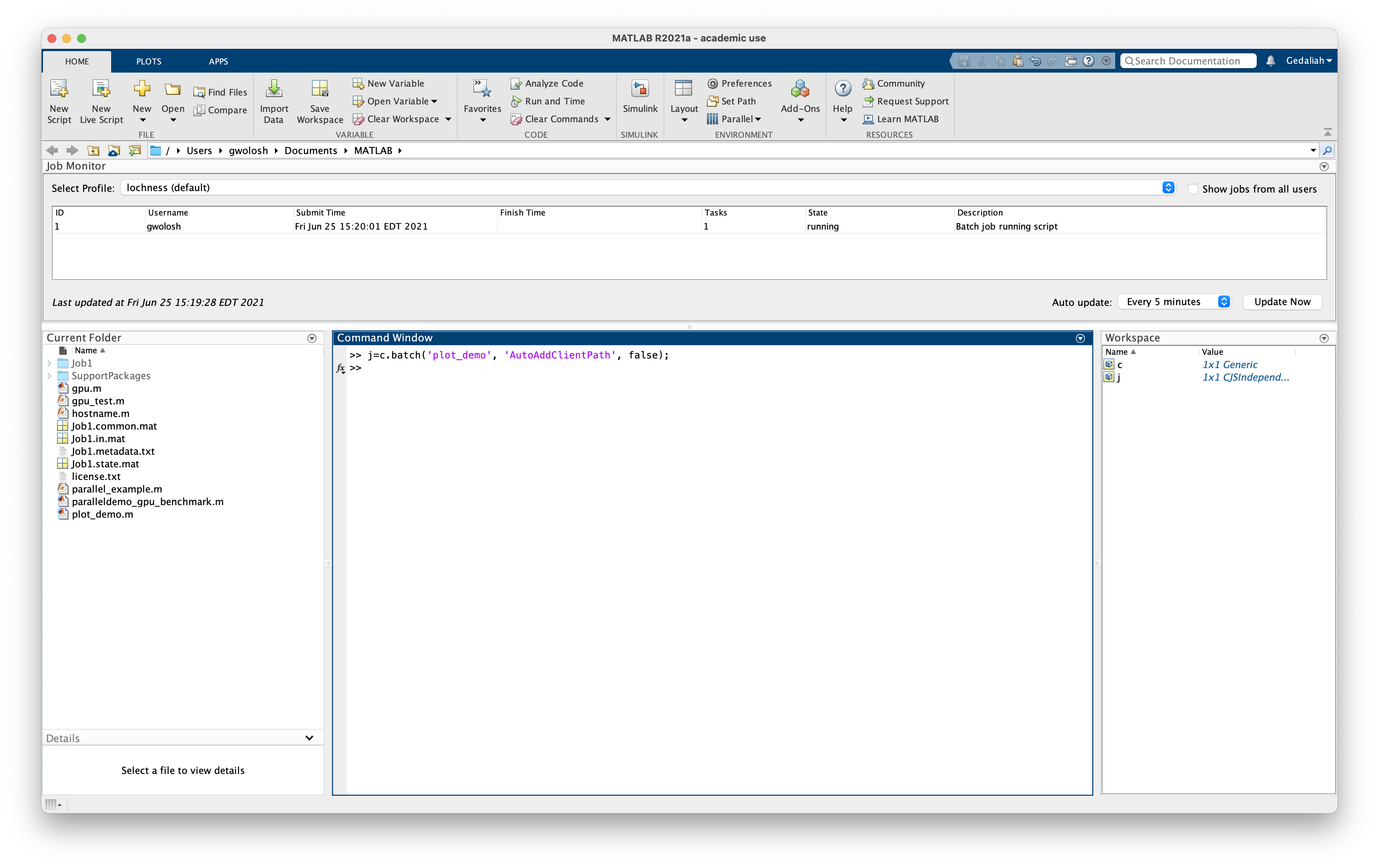{ width=70% height 70%} - -To load 'x' into the local Matlab workspace: -``` - >> load(j,'x') -``` - { width=70% height 70%} - - Finally, plot the results: -``` - >> plot(x) -``` -{ width=70% height 70%} - \ No newline at end of file diff --git a/docs/Software/md/gromacs.md b/docs/Software/md/gromacs.md deleted file mode 100644 index 9b9bdf60f..000000000 --- a/docs/Software/md/gromacs.md +++ /dev/null @@ -1,42 +0,0 @@ ---- -title: GROMACS ---- - -[GROMACS](https://www.gromacs.org) is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. - -It is primarily designed for biochemical molecules like proteins, lipids and nucleic acids that have a lot of complicated bonded interactions, but since GROMACS is extremely fast at calculating the nonbonded interactions (that usually dominate simulations) many groups are also using it for research on non-biological systems, e.g. polymers. - -## Availability - -=== "Wulver" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - soft = df.query('Software == "GROMACS"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - soft = df.query('Software == "GROMACS"') - print(soft.to_markdown(index=False)) - ``` - -## Related Applications - -* [LAMMPS](lammps.md) - -## User Contributed Information - -!!! info "Please help us improve this page" - - Users are invited to contribute helpful information and corrections through our [Github repository](https://github.com/arcs-njit-edu/Docs/blob/main/CONTRIBUTING.md). - - - diff --git a/docs/Software/programming/compilers.md b/docs/Software/programming/compilers.md index 4a8ee7a92..59b2f3572 100644 --- a/docs/Software/programming/compilers.md +++ b/docs/Software/programming/compilers.md @@ -10,21 +10,7 @@ We offer both GNU and Intel compilers. Here is the list of compilers you can fin ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - # Header values to be added - #column_names=["Compilers","modules"] - soft = df.query('Software == "GCC" | Software == "intel-compilers"') - soft = soft[~soft.apply(lambda row: row.astype(str).str.contains('system').any(), axis=1)] - - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') # Header values to be added #column_names=["Compilers","modules"] soft = df.query('Software == "GCC" | Software == "intel-compilers"') @@ -40,22 +26,13 @@ MPI (Message Passing Interface) libraries are a set of software tools that allow ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') soft = df.query('Software == "OpenMPI" | Software == "impi"') #soft.columns = column_names print(soft.to_markdown(index=False)) ``` -=== "Lochness" - ```python exec="on" - import pandas as pd - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - - soft = df.query('Software == "OpenMPI" | Software == "impi"') - #soft.columns = column_names - print(soft.to_markdown(index=False)) - ``` ## Toolchains We use [EasyBuild](https://easybuild.io) to install the packages as modules and to avoid too many packages tol load as a module, we use pre-defined build environment modules called toolchains which include a combination of tools such as compilers, libraries etc. We use `foss` and `intel` toolchains in Wulver. The advantage of using toolchains is that user can load either `foss` or `intel` as base package and the additional libraries such as MPI, LAPACK and other math libraries will be automatically loaded. @@ -91,20 +68,7 @@ foss ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - # Header values to be added - #column_names=["Toolchains","modules"] - soft = df.query('Software == "foss"') - #soft.columns = column_names - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') # Header values to be added #column_names=["Toolchains","modules"] soft = df.query('Software == "foss"') @@ -135,20 +99,7 @@ intel ```python exec="on" import pandas as pd - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - # Header values to be added - #column_names=["Toolchains","modules"] - soft = df.query('Software == "intel"') - #soft.columns = column_names - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') + df = pd.read_csv('docs/assets/tables/module_wulver_rhel9.csv') # Header values to be added #column_names=["Toolchains","modules"] soft = df.query('Software == "intel"') diff --git a/docs/Software/programming/python/conda.md b/docs/Software/programming/python/conda.md deleted file mode 100644 index e40c67196..000000000 --- a/docs/Software/programming/python/conda.md +++ /dev/null @@ -1,370 +0,0 @@ -Since Python supports a wide a range additional libraries in machine learning or datas science research, it is not always possible to install every package on HPC. Also, users sometimes need to use a specific version of Python or its libraries to conduct their research. Therefore, in that case users can build their own Python version along with specific library. One of the way to accomplish this is to use Conda. - -# Conda -Conda as a package manager helps you find and install packages. If you need a package that requires a different version of Python, you do not need to switch to different environment manager, because conda is also an environment manager. - -## Availability -Conda can be accessed on cluster as `Anaconda3` or `Miniconda3` module. - -=== "Wulver" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_wulver.csv') - # Header values to be added - soft = df.query('Software == "Anaconda3" | Software == "Miniconda3"') - print(soft.to_markdown(index=False)) - ``` - -=== "Lochness" - - ```python exec="on" - import pandas as pd - - df = pd.read_csv('docs/assets/tables/module_lochness.csv') - # Header values to be added - soft = df.query('Software == "Anaconda3" | Software == "Miniconda3"') - print(soft.to_markdown(index=False)) - ``` - -Users can use conda after using any of the modules mentioned above - -module a `Anaconda3` module. Users can use `Anaconda3` to create virtual python environments to manage python modules. - -## Create and Activate a Conda Virtual Environment - -Load the miniconda Module - -``` -module load Anaconda3 -``` - -### Create Environment with `conda` -To create an environment use the `conda create` command. Once the environment is created you need to create a file on `$HOME` directory and add the following - -??? Example "conda3.sh" - - ```bash - # >>> conda initialize >>> - # !! Contents within this block are managed by 'conda init' !! - __conda_setup="$('conda' 'shell.bash' 'hook' 2> /dev/null)" - if [ $? -eq 0 ]; then - eval "$__conda_setup" - else - if [ -f "$EBROOTANACONDA3/etc/profile.d/conda.sh" ]; then - . "$EBROOTANACONDA3/etc/profile.d/conda.sh" - else - export PATH="$EBROOTANACONDA3/bin:$PATH" - fi - fi - unset __conda_setup - # <<< conda initialize <<< - ``` -Before activating the environment You need to use `source ~/conda3.sh` to activate the path. - -### Examples -Here, we provide instructions on how to use `conda` to install application - -#### Install TensorFlow with GPU -The following example will create a new conda environment based on python 3.9 and install tensorflow in the environment. - -```bash -login1-41 ~ >: module load Anaconda3 -login1-41 ~ >: conda create --name tf python=3.9 -Collecting package metadata (current_repodata.json): done -Solving environment: done - -## Package Plan ## - - environment location: /home/g/guest24/.conda/envs/tf - - added / updated specs: - - python=3.9 - - -The following packages will be downloaded: - -