
@@ -94,6 +88,10 @@ Alternatively, you can specify known (model, dataset) pairs using the `--model-d
tti-eval evaluate --model-dataset clip/Alzheimer-MRI --model-dataset bioclip/Alzheimer-MRI
```
+The evaluation results can be exported to a CSV file using the `--save` option.
+They will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, exported evaluation results are stored to the `output/evaluations` folder within the repository.
+
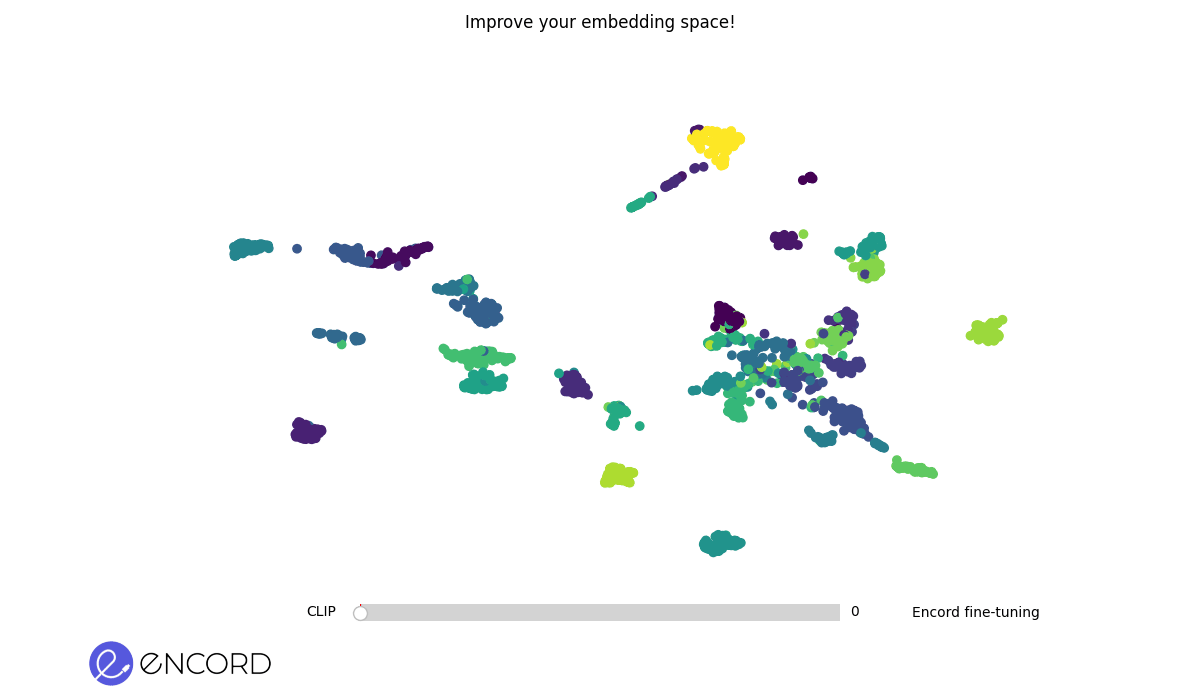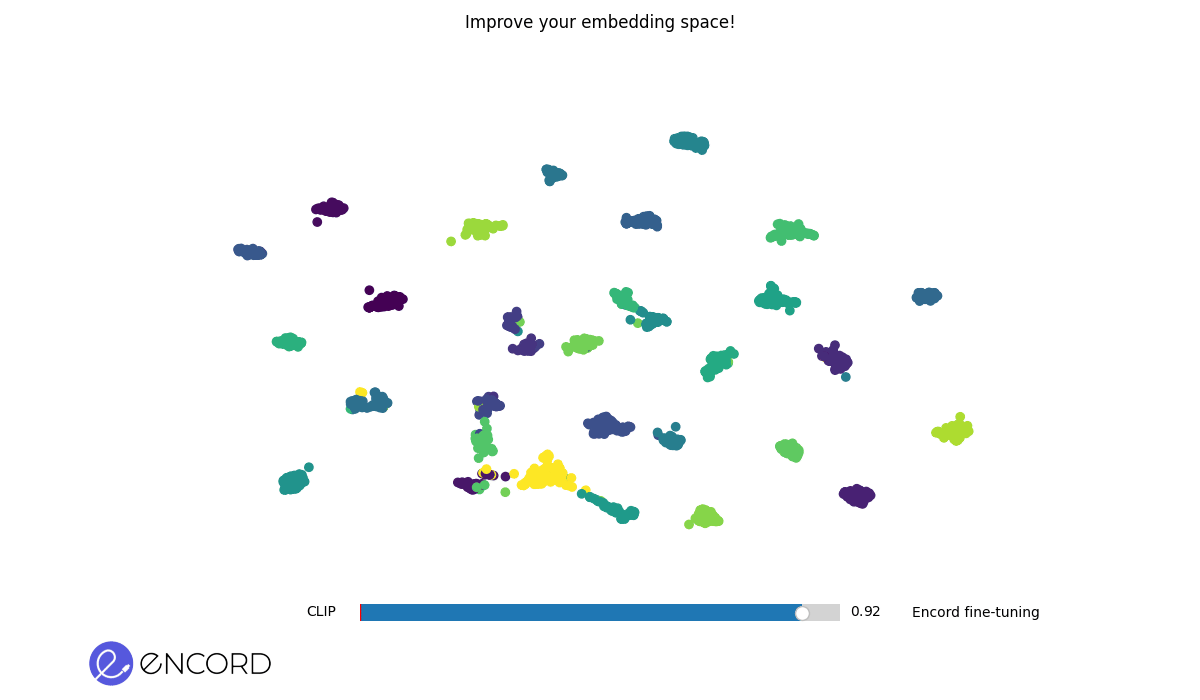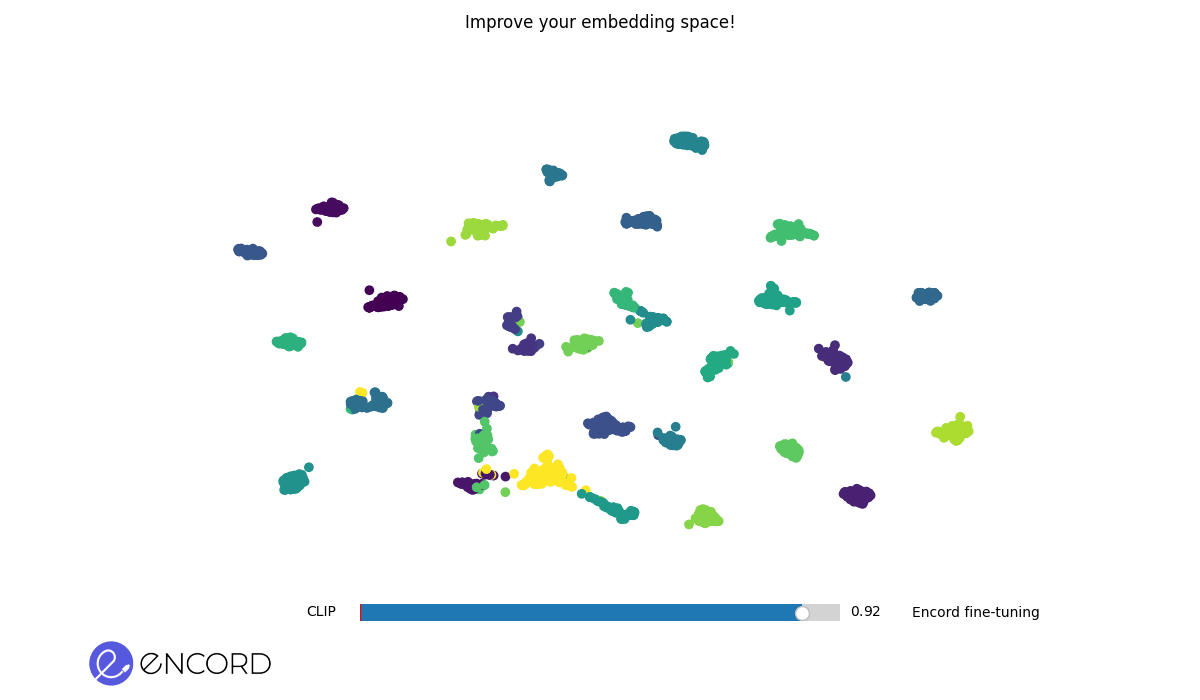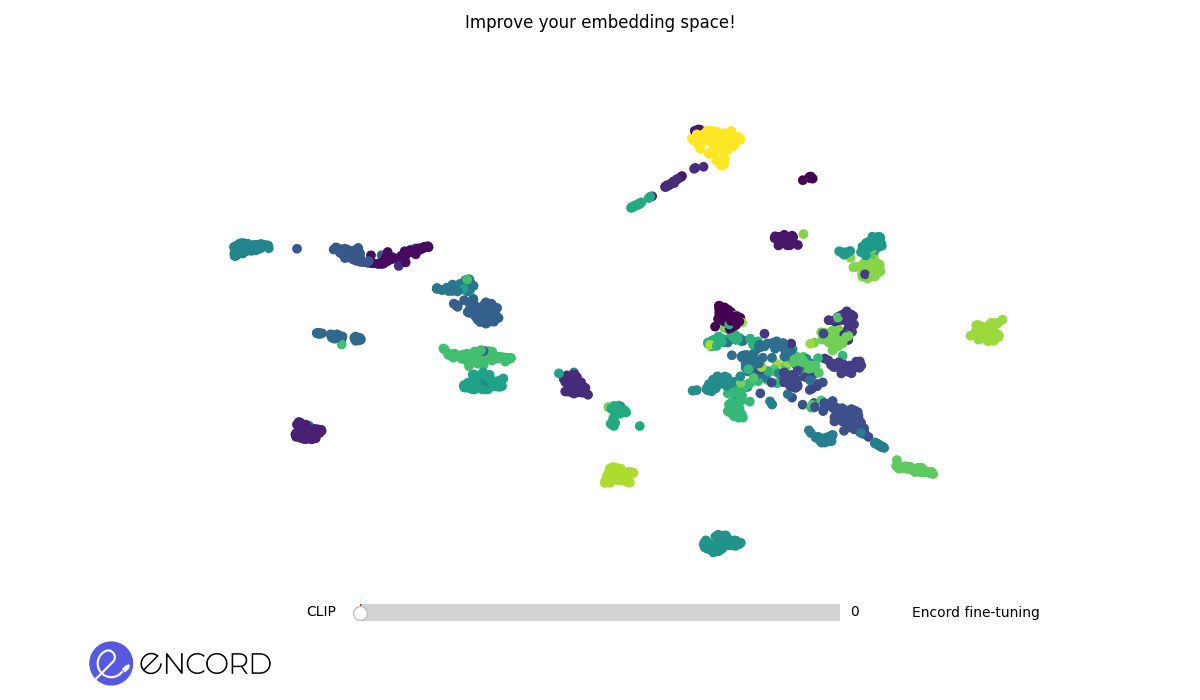
### Embeddings Animation
To create 2D animations of the embeddings, use the CLI command `tti-eval animate`.
@@ -106,12 +104,12 @@ Alternatively, you can specify the models and dataset as arguments. For example:
tti-eval animate clip bioclip Alzheimer-MRI
```
-The animations will be saved at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
-By default, this path corresponds to the `output` folder in the repository directory.
+The animations will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, animations are stored in the `output/animations` folder within the repository.
To interactively explore the animation in a temporary session, use the `--interactive` flag.
-
+

> ℹ️ You can also carry out these operations using Python. Explore our Python Quickstart guide for more details.
 -
-
-
- -
-
-
- @@ -94,6 +88,10 @@ Alternatively, you can specify known (model, dataset) pairs using the `--model-d
tti-eval evaluate --model-dataset clip/Alzheimer-MRI --model-dataset bioclip/Alzheimer-MRI
```
+The evaluation results can be exported to a CSV file using the `--save` option.
+They will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, exported evaluation results are stored to the `output/evaluations` folder within the repository.
+
### Embeddings Animation
To create 2D animations of the embeddings, use the CLI command `tti-eval animate`.
@@ -106,12 +104,12 @@ Alternatively, you can specify the models and dataset as arguments. For example:
tti-eval animate clip bioclip Alzheimer-MRI
```
-The animations will be saved at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
-By default, this path corresponds to the `output` folder in the repository directory.
+The animations will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, animations are stored in the `output/animations` folder within the repository.
To interactively explore the animation in a temporary session, use the `--interactive` flag.
@@ -94,6 +88,10 @@ Alternatively, you can specify known (model, dataset) pairs using the `--model-d
tti-eval evaluate --model-dataset clip/Alzheimer-MRI --model-dataset bioclip/Alzheimer-MRI
```
+The evaluation results can be exported to a CSV file using the `--save` option.
+They will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, exported evaluation results are stored to the `output/evaluations` folder within the repository.
+
### Embeddings Animation
To create 2D animations of the embeddings, use the CLI command `tti-eval animate`.
@@ -106,12 +104,12 @@ Alternatively, you can specify the models and dataset as arguments. For example:
tti-eval animate clip bioclip Alzheimer-MRI
```
-The animations will be saved at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
-By default, this path corresponds to the `output` folder in the repository directory.
+The animations will be saved on a folder at the location specified by the environment variable `TTI_EVAL_OUTPUT_PATH`.
+By default, animations are stored in the `output/animations` folder within the repository.
To interactively explore the animation in a temporary session, use the `--interactive` flag.
 +
+ 