 +
+
+
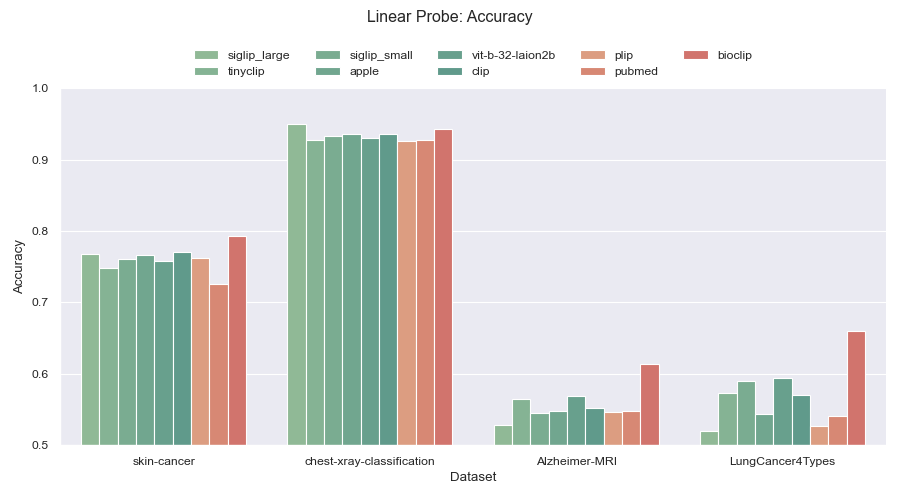
+Figure 1: Linear probe accuracy across four different medical datasets. General purpose models are colored green while models trained for the medical domain are colored red. +
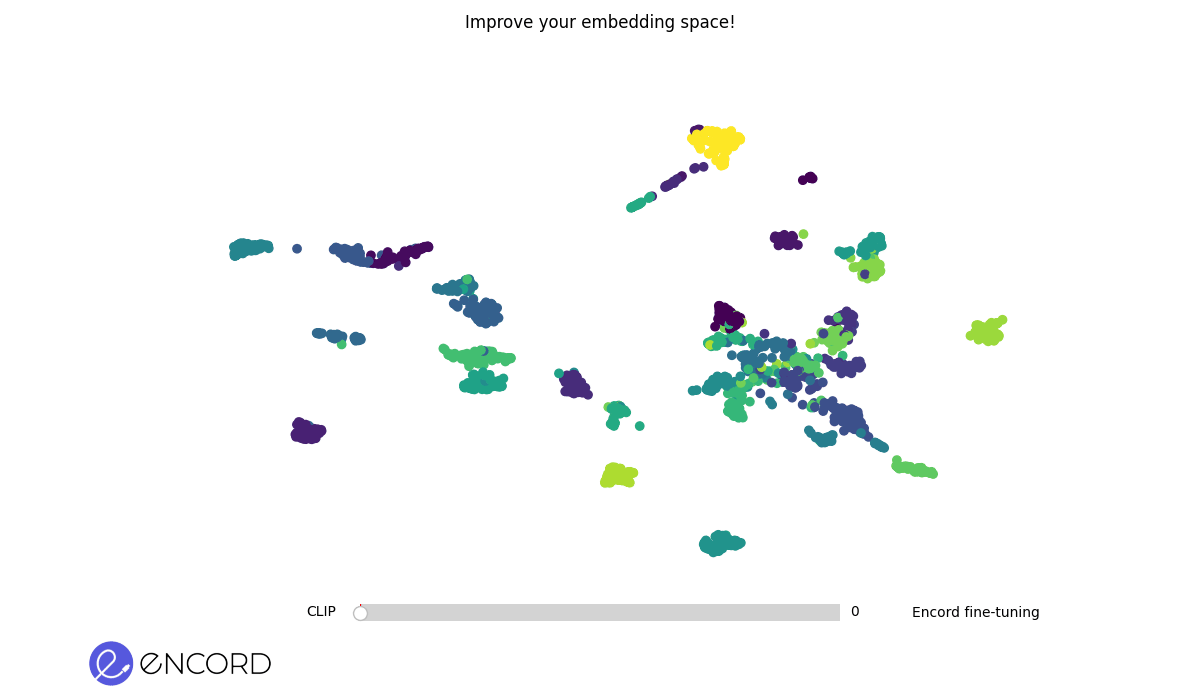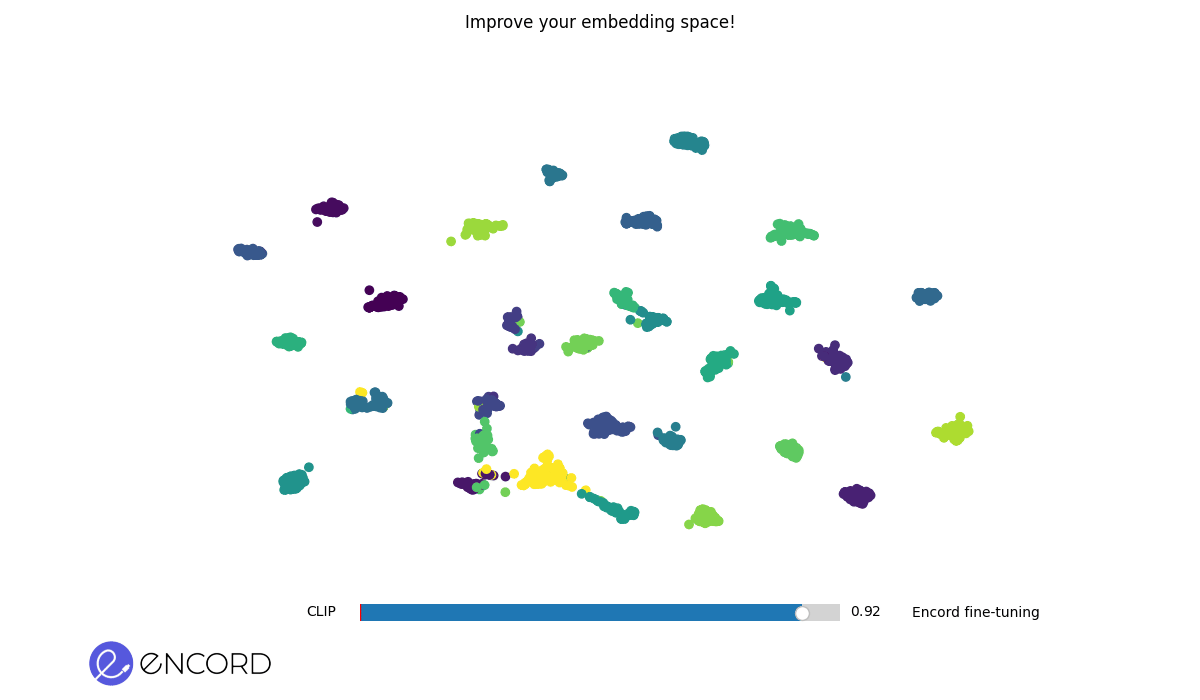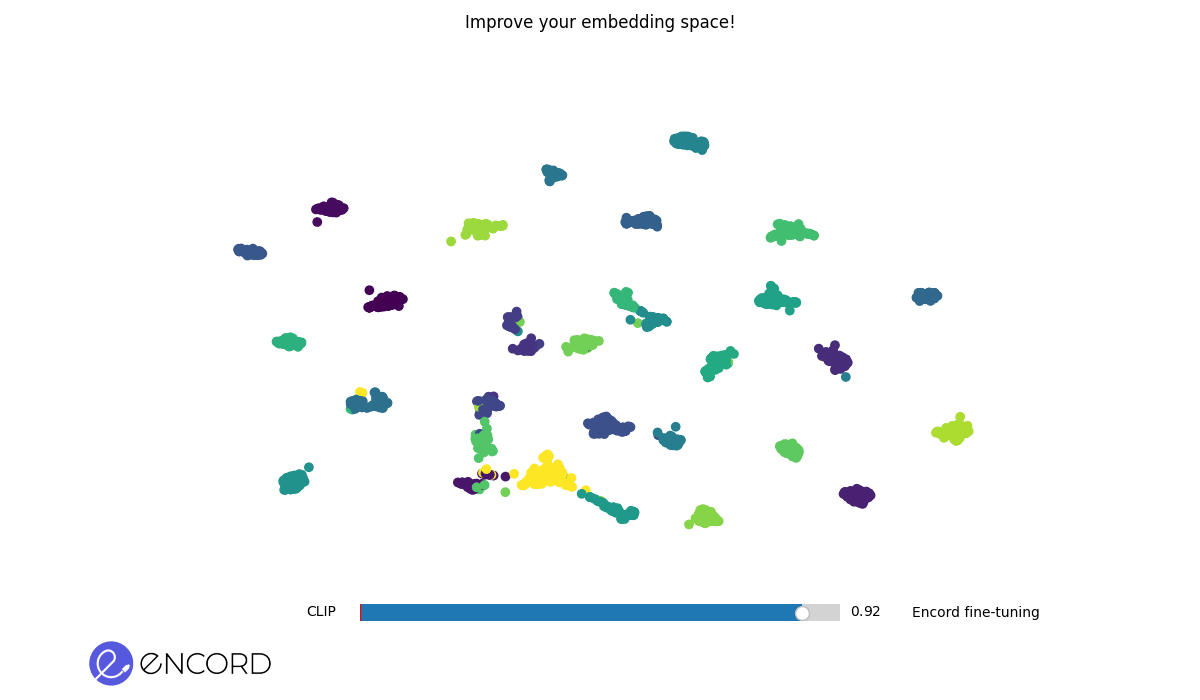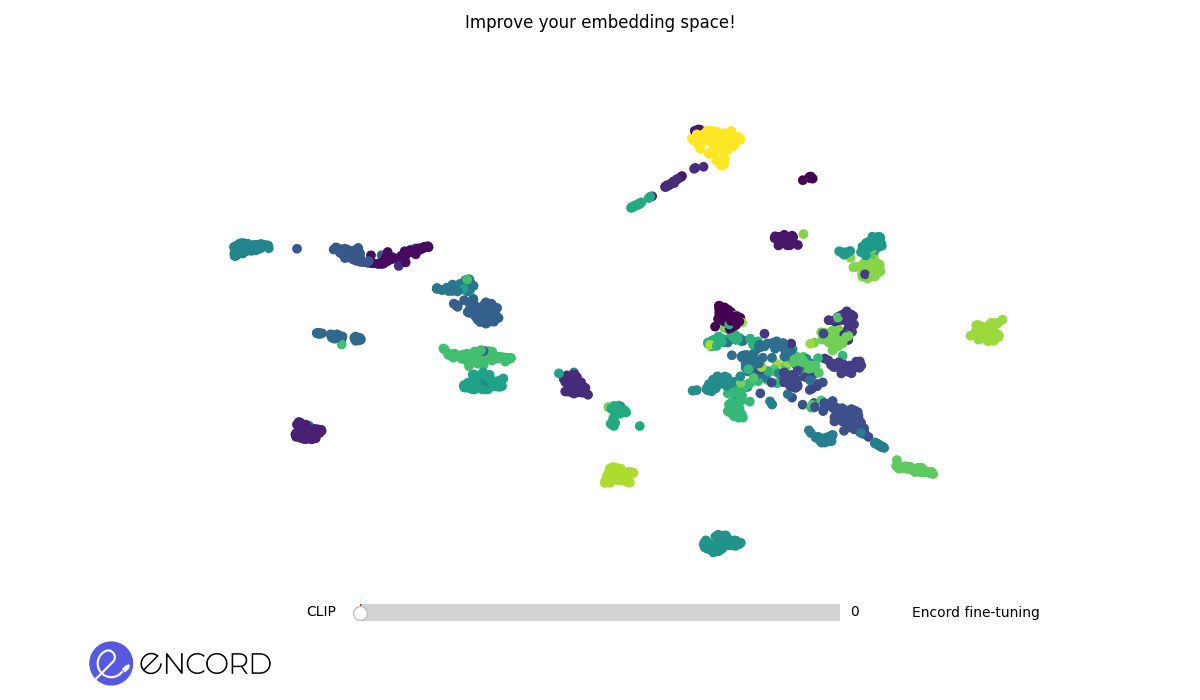 +## Some Example Results
+
+One example of where this `tti-eval` is useful is to test different open-source models against different open-source datasets within a specific domain.
+Below, we focused on the medical domain. We evaluate nine different models of which three of them are domain specific.
+The models are evaluated against four different medical datasets. Note, Further down this page, you will find links to all models and datasets.
+
+
+## Some Example Results
+
+One example of where this `tti-eval` is useful is to test different open-source models against different open-source datasets within a specific domain.
+Below, we focused on the medical domain. We evaluate nine different models of which three of them are domain specific.
+The models are evaluated against four different medical datasets. Note, Further down this page, you will find links to all models and datasets.
+
+ +
+ Figure 1: Linear probe accuracy across four different medical datasets. General purpose models are colored green while models trained for the medical domain are colored red. +