CAFE (Computational Analysis of gene Family Evolution) is a software to analyze changes in gene family size in a way that accounts for phylogenetic history and provides a statistical foundation for evolutionary inferences.
CAFE5 currently does not provide tools to visualize Expansion/Contraction of gene families on phylogenetic tree. To solve this issue, I developed CafePlotter, a tool for plotting CAFE5 gene family expansion/contraction result.
Python 3.8 or later is required for installation.
Install PyPI package:
pip install cafeplotter
cafeplotter -i [CAFE5 result directory] -o [Output directory]
General Options:
-i IN, --indir IN CAFE5 result directory as input
-o OUT, --outdir OUT Output directory for plotting CAFE5 result
--format Output image format ('png'[default]|'jpg'|'svg'|'pdf')
-v, --version Print version information
-h, --help Show this help message and exit
Figure Appearence Options:
--fig_height Figure height per leaf node of tree (Default: 0.5)
--fig_width Figure width (Default: 8.0)
--leaf_label_size Leaf label size (Default: 12)
--count_label_size Gene count label size (Default: 8)
--innode_label_size Internal node label size (Default: 0)
--p_label_size Branch p-value label size (Default: 0)
--ignore_branch_length Ignore branch length for plotting tree (Default: OFF)
--expansion_color Plot color of gene family expansion (Default: 'red')
--contraction_color Plot color of gene family contraction (Default: 'blue')
--dpi Figure DPI (Default: 300)
User can download example dataset (singlelambda.zip):
cafeplotter -i ./examples/singlelambda -o ./singlelambda_plot --ignore_branch_length
-
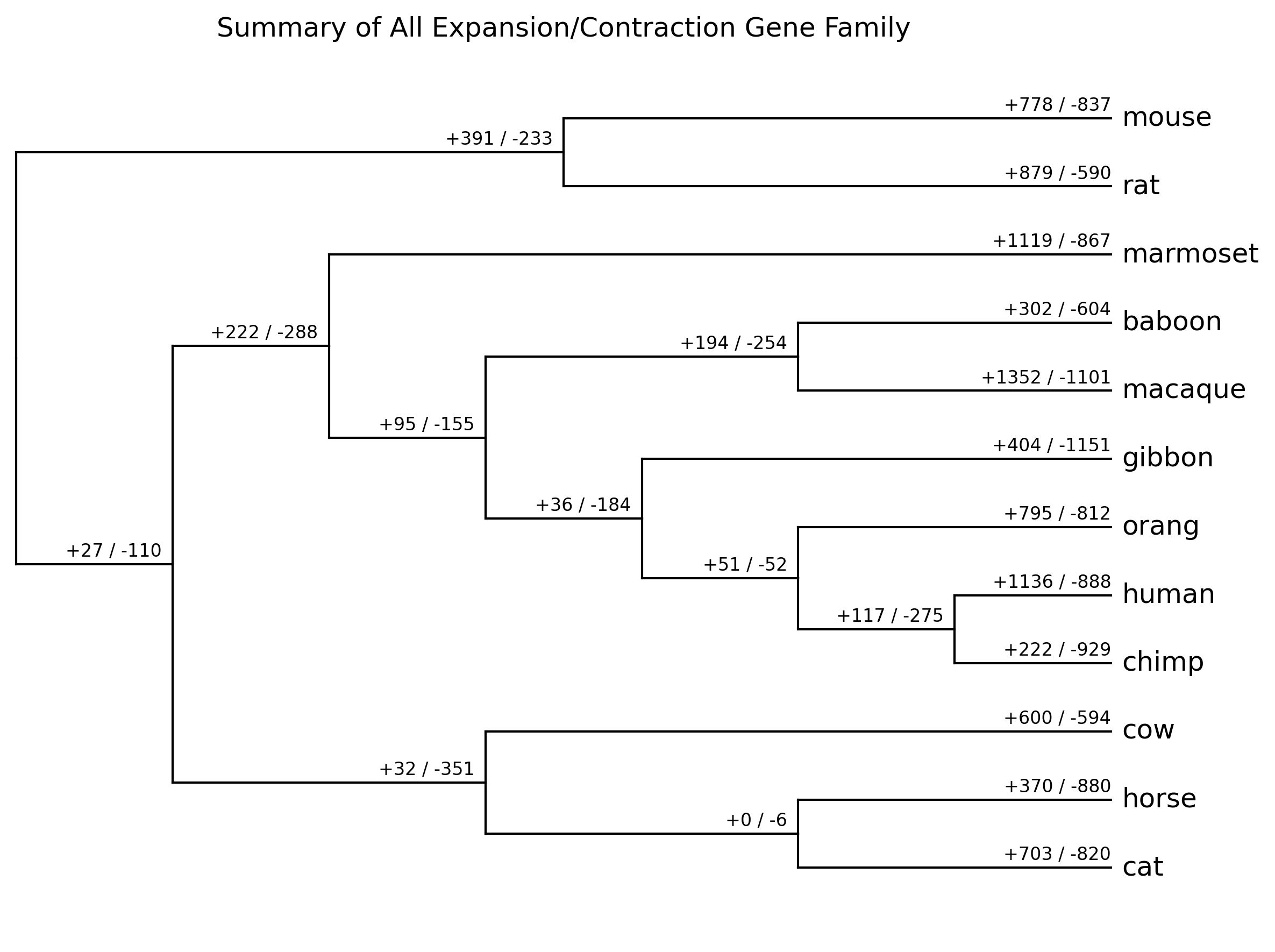
summary_all_gene_family.[png|jpg|svg|pdf]
Summary of all expansion/contraction gene family result (from*_clade_result.txt) -
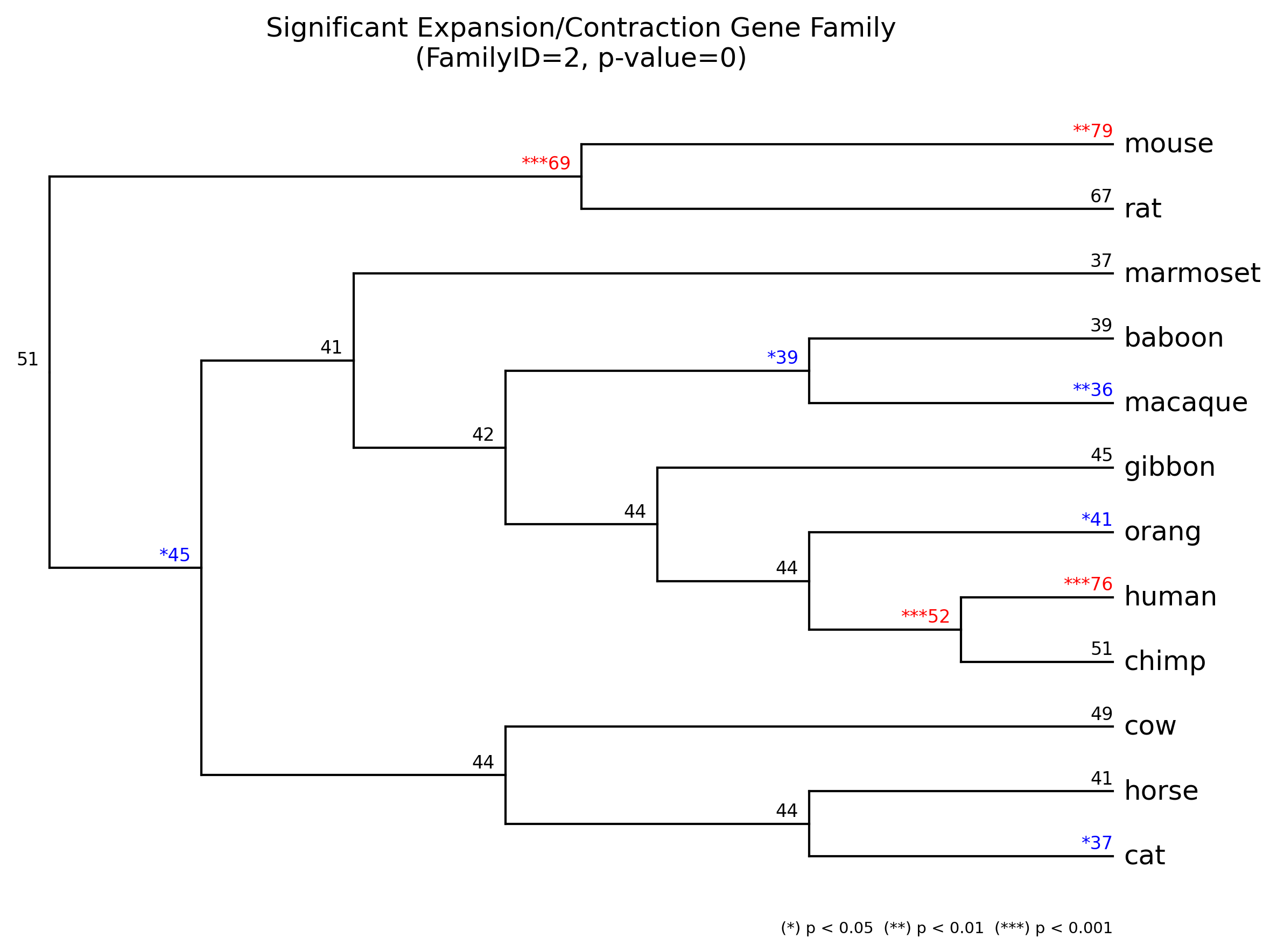
gene_family/{FamilyID}_gene_family.[png|jpg|svg|pdf]
Significant expansion(red)/contraction(blue) gene family result -
result_summary.tsv (example)
Significant expansion/contraction result summary for each family and taxon