This tutorial is an introduction to visualising and analysing spatial data in R based on the sp class system. For a guide to the more recent sf package check out Chapter 2 of the in-development book Geocomputation with R, the source code of which can be found at github.com/Robinlovelace/geocompr.
Although sf supersedes sp in many ways, there is still merit in learning the content in this tutorial, which teaches principles that will be useful regardless of software. Specifically this tutorial focusses on map-making with R's 'base' graphics and various dedicated map-making packages for R including tmap and leaflet. It aims to teach the basics of using R as a fast, user-friendly and extremely powerful command-line Geographic Information System (GIS).
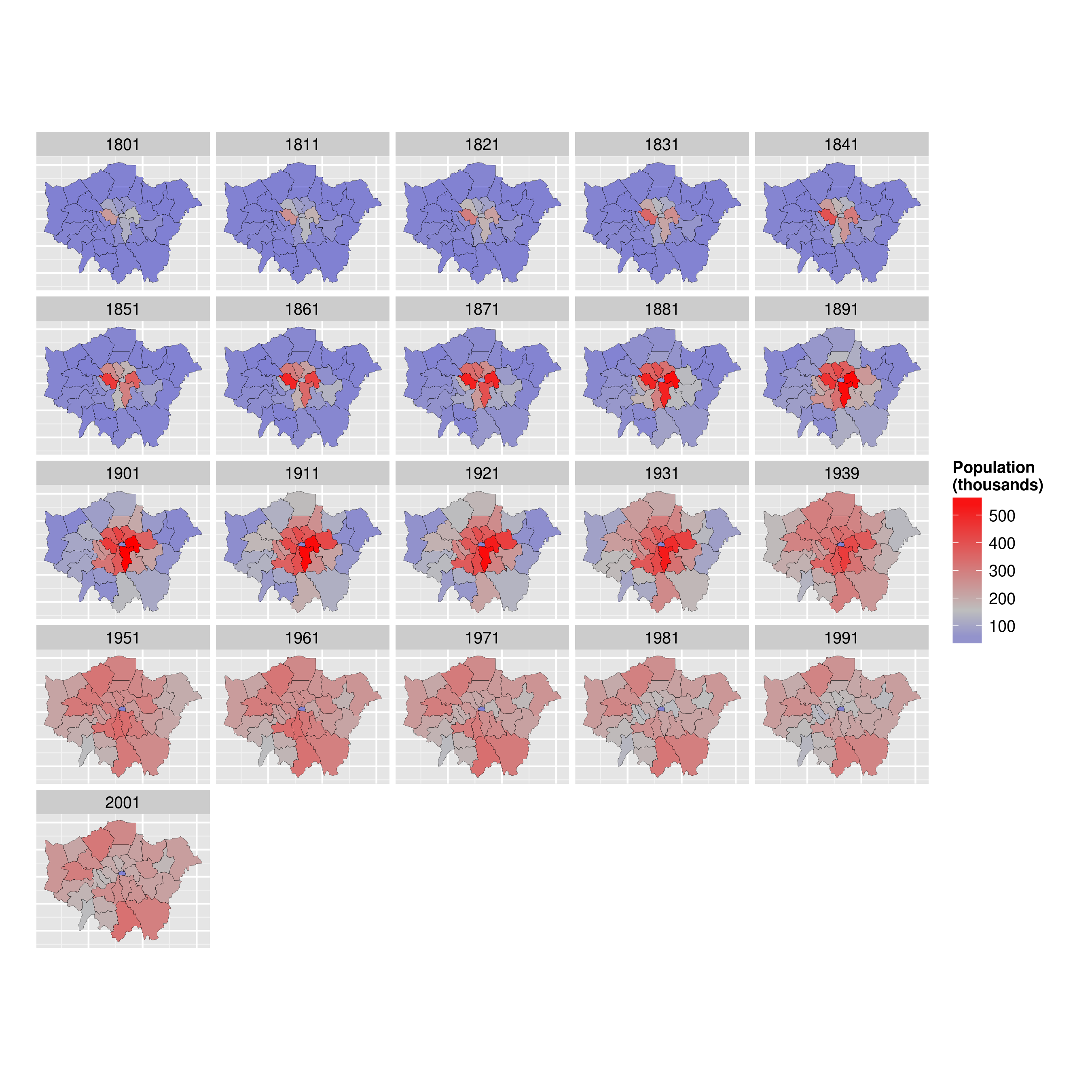
By the end of the tutorial you should have the confidence and skills needed to convert a diverse range of geographical and non-geographical datasets into meaningful analyses and visualisations. Using data and code provided in this repository all of the results are reproducible, culminating in publication-quality maps such as the faceted map of London's population below:
The course will even show you how to make your maps animated:
An up-to-date pdf version of this tutorial is maintained for teaching purposes in the file intro-spatial-rl.pdf.
If you have any feedback on this tutorial please let us know via email or via this repository. Contibutions to the .Rmd file (README.Rmd) are welcome. Happy mapping!
The tutorial is practical in nature: you will load-in, visualise and manipulate spatial data. We assume no prior knowledge of spatial data analysis but some experience with R will help. If you have not used R before, it may be worth following an introductory tutorial, such as Efficient R Programming (Gillespie and Lovelace, 2016), R for Data Science (Grolemund and Wickham, 2016) or tutorials suggested on rstudio.com and cran.r-project.org.
...
Now you know some R, it's time to turn your attention towards spatial data with R. To that end, this tutorial is organised as follows:
- Introduction: provides a guide to R's syntax and preparing for the tutorial
- Spatial data in R: describes basic spatial functions in R
- Creating and manipulating spatial data: includes changing projection, clipping and spatial joins
- Map making with tmap, ggplot2 and leaflet: this section demonstrates map making with more advanced visualisation tools
- Taking spatial analysis in R further: a compilation of resources for furthering your skills
To distinguish between prose and code, please be aware of the following typographic conventions used in this document: R code (e.g. plot(x, y)) is written in a monospace font and package names (e.g. rgdal) are written in bold. A double hash (##) at the start of a line of code indicates that this is output from R. Lengthy outputs have been omitted from the document to save space, so do not be alarmed if R produces additional messages: you can always look up them up on-line.
As with any programming language, there are often many ways to produce the same output in R. The code presented in this document is not the only way to do things. We encourage you to play with the code to gain a deeper understanding of R. Do not worry, you cannot 'break' anything using R and all the input data can be re-loaded if things do go wrong. As with learning to skateboard, you learn by falling and getting an Error: message in R is much less painful than falling onto concrete! We encourage Error:s --- it means you are trying new things.
For this tutorial you need a copy of R. The latest version can be downloaded from http://cran.r-project.org/.
We also suggest that you use an R editor, such as RStudio, as this will improve the user-experience and help with the learning process. This can be downloaded from http://www.rstudio.com. The R Studio interface is comprised of a number of windows, the most important being the console window and the script window. Anything you type directly into the console window will not be saved, so use the script window to create scripts which you can save for later use. There is also a Data Environment window which lists the dataframes and objects being used. Familiarise yourself with the R Studio interface before getting started on the tutorial.
When writing code in any language, it is good practice to use consistent and clear conventions, and R is no exception. Adding comments to your code is also useful; make these meaningful so you remember what the code is doing when you revisit it at a later date. You can add a comment by using the # symbol before or after a line of code, as illustrated in the block of code below. This code should create Figure 1 if typed correctly into the Console window:
This first line in this block of code creates a new object called x and assigns it to a range of integers between 1 and 400. The second line creates another object called y which is assigned to a mathematical formula, and the third line plots the two together to create the plot shown.
Note <-, the directional "arrow" assignment symbol which creates a new object and assigns it to the value you have given.[1]
If you require help on any function, use the help command, e.g. help(plot). Because R users love being concise, this can also be written as ?plot. Feel free to use it at any point you would like more detail on a specific function (although R's help files are famously cryptic for the un-initiated). Help on more general terms can be found using the ?? symbol. To test this, try typing ??regression. For the most part, learning by doing is a good motto, so let's crack on and download some packages and data.
R has a huge and growing number of spatial data packages. We recommend taking a quick browse on R's main website to see the spatial packages available: http://cran.r-project.org/web/views/Spatial.html.
In this tutorial we will use the packages from the 'spverse', that use the sp package:
- ggmap: extends the plotting package ggplot2 for maps
- rgdal: R's interface to the popular C/C++ spatial data processing library gdal
- rgeos: R's interface to the powerful vector processing library geos
- maptools: provides various mapping functions
- dplyr and tidyr: fast and concise data manipulation packages
- tmap: a new packages for rapidly creating beautiful maps
For a tutorial based on the recent sf package you will have to look elsewhere, like the geocompr website, the online home of the forthcoming book Geocomputation with R.
Some packages may already be installed on your computer. To test if a package is installed, try to load it using the library function; for example, to test if ggplot2 is installed, type library(ggplot2) into the console window. If there is no output from R, this is good news: it means that the library has already been installed on your computer.
If you get an error message,you will need to install the package using install.packages("ggplot2"). The package will download from the Comprehensive R Archive Network (CRAN); if you are prompted to select a 'mirror', select one that is close to current location. If you have not done so already, install these packages on your computer now. A quick way to do this in one go is to enter the following lines of code:
x <- c("ggmap", "rgdal", "rgeos", "maptools", "dplyr", "tidyr", "tmap")
# install.packages(x) # warning: uncommenting this may take a number of minutes
lapply(x, library, character.only = TRUE) # load the required packagesNow that we have looked at R's basic syntax and installed the necessary packages,let's load some real spatial data. The next part of the tutorial will focus on plotting and interrogating spatial objects.
The data used for this tutorial can be downloaded from: https://github.com/Robinlovelace/Creating-maps-in-R. Click on the "Download ZIP" button on the right hand side of the screen and once downloaded, unzip this to a new folder on your computer.
Open the existing 'Creating-maps-in-R' project using File -> Open File... on the top menu.
Alternatively, use the project menu to open the project or create a new one. It is highly recommended that you use RStudio's projects to organise your R work and that you organise your files into sub-folders (e.g. code, input-data, figures) to avoid digital clutter (Figure 2). The RStudio website contains an overview of the software: rstudio.com/products/rstudio/.
Opening a project sets the current working directory to the project's parent folder, the Creating-maps-in-R folder in this case. If you ever need to change your working directory, you can use the 'Session' menu at the top of the page or use the setwd command.
The first file we are going to load into R Studio is the "london_sport" shapefile located in the 'data' folder of the project. It is worth looking at this input dataset in your file browser before opening it in R. You will notice that there are several files named "london_sport", all with different file extensions. This is because a shapefile is actually made up of a number of different files, such as .prj, .dbf and .shp.
You could also try opening the file "london_sport.shp" file in a conventional GIS such as QGIS to see what a shapefile contains.
You should also open "london_sport.dbf" in a spreadsheet program such as LibreOffice Calc. to see what this file contains. Once you think you understand the input data, it's time to open it in R. There are a number of ways to do this, the most commonly used and versatile of which is readOGR. This function, from the rgdal package, automatically extracts the information regarding the data.
rgdal is R’s interface to the "Geospatial Abstraction Library (GDAL)" which is used by other open source GIS packages such as QGIS and enables R to handle a broader range of spatial data formats. If you've not already installed and loaded the rgdal package (see the 'prerequisites and packages' section) do so now:
library(rgdal)
lnd <- readOGR(dsn = "data/london_sport.shp")
# lnd <- readOGR(dsn = "data", layer = "london_sport")In the second line of code above the readOGR function is used to load a shapefile and assign it to a new spatial object called lnd, short for London.
readOGR is a function of the rgdal package, the first argument of which dsn: "data source name", the file or directory of the geographic data to be loaded. Thanks to recent developments in rgdal, the layer no longer has to be specified (as it is in the 3 line of code which has been commented out).[2]
lnd is now an object representing the population of London Boroughs in 2001 and the percentage of the population participating in sporting activities according to the Active People Survey. The boundary data is from the Ordnance Survey.
For information about how to load different types of spatial data, see the help documentation for readOGR. This can be accessed by typing ?readOGR. For another worked example, in which a GPS trace is loaded, please see Cheshire and Lovelace (2014).
Spatial objects like the lnd object are made up of a number of different slots, the key slots being @data (non geographic attribute data) and @polygons (or @lines for line data). The data slot can be thought of as an attribute table and the geometry slot is the polygons that make up the physcial boundaries. Specific slots are accessed using the @ symbol. Let's now analyse the sport object with some basic commands:
head(lnd@data, n = 2)## ons_label name Partic_Per Pop_2001
## 0 00AF Bromley 21.7 295535
## 1 00BD Richmond upon Thames 26.6 172330
mean(lnd$Partic_Per) # short for mean(lnd@data$Partic_Per) ## [1] 20.05455
Take a look at the output created (note the table format of the data and the column names). There are two important symbols at work in the above block of code: the @ symbol in the first line of code is used to refer to the data slot of the lnd object. The $ symbol refers to the Partic_Per column (a variable within the table) in the data slot, which was identified from the result of running the first line of code.
The head function in the first line of the code above simply means "show the first few lines of data" (try entering head(lnd@data), see ?head for more details). The second line calculates finds the mean sports participation per 100 people for zones in London. The results works because we are dealing with numeric data. To check the classes of all the variables in a spatial dataset, you can use the following command:
sapply(lnd@data, class)## ons_label name Partic_Per Pop_2001
## "factor" "factor" "numeric" "factor"
This shows that, unexpectedly, Pop_2001 is a factor. We can coerce the variable into the correct, numeric, format with the following command:
lnd$Pop_2001 <- as.numeric(as.character(lnd$Pop_2001))Type the function again but this time hit tab before completing the command. RStudio has auto-complete functionality which can save you a lot of time in the long run (see Figure 3).
To explore lnd object further, try typing nrow(lnd) (display number of rows) and record how many zones the dataset contains. You can also try ncol(lnd).
One issue with the data that we have loaded is that it has no coordinate reference system (CRS):
lnd@proj4string## CRS arguments:
## +proj=tmerc +lat_0=49 +lon_0=-2 +k=0.9996012717 +x_0=400000
## +y_0=-100000 +ellps=airy +units=m +no_defs
Now we have seen something of the structure of spatial objects in R, let us look at plotting them. Note, that plots use the geometry data, contained primarily in the @polygons slot.
plot(lnd) # not shown in tutorial - try it on your computerplot is one of the most useful functions in R, as it changes its behaviour depending on the input data (this is called polymorphism by computer scientists). Inputting another object such as plot(lnd@data) will generate an entirely different type of plot. Thus R is intelligent at guessing what you want to do with the data you provide it with.
R has powerful subsetting capabilities that can be accessed very concisely using square brackets,as shown in the following example:
# select rows of lnd@data where sports participation is less than 13
lnd@data[lnd$Partic_Per < 13, 1:3]## ons_label name Partic_Per
## 32 00AA City of London 9.1
The above line of code asked R to select only the rows from the lnd object, where sports participation is lower than 13, in this case rows 17, 21 and 32, which are Harrow, Newham and the city centre respectively. The square brackets work as follows: anything before the comma refers to the rows that will be selected, anything after the comma refers to the number of columns that should be returned. For example if the data frame had 1000 columns and you were only interested in the first two columns you could specify 1:2 after the comma. The ":" symbol simply means "to", i.e. columns 1 to 2. Try experimenting with the square brackets notation (e.g. guess the result of lnd@data[1:2, 1:3] and test it).
So far we have been interrogating only the attribute data slot (@data) of the lnd object, but the square brackets can also be used to subset spatial objects, i.e. the geometry slot. Using the same logic as before try to plot a subset of zones with high sports participation.
# Select zones where sports participation is between 20 and 25%
sel <- lnd$Partic_Per > 20 & lnd$Partic_Per < 25
plot(lnd[sel, ]) # output not shown here
head(sel) # test output of previous selection (not shown)This plot is quite useful, but it only displays the areas which meet the criteria. To see the sporty areas in context with the other areas of the map simply use the add = TRUE argument after the initial plot. (add = T would also work, but we like to spell things out in this tutorial for clarity). What do you think the col argument refers to in the below block? (see Figure 5).
If you wish to experiment with multiple criteria queries, use &.
plot(lnd, col = "lightgrey") # plot the london_sport object
sel <- lnd$Partic_Per > 25
plot(lnd[ sel, ], col = "turquoise", add = TRUE) # add selected zones to mapCongratulations! You have just interrogated and visualised a spatial object: where are areas with high levels of sports participation in London? The map tells us. Do not worry for now about the intricacies of how this was achieved: you have learned vital basics of how R works as a language; we will cover this in more detail in subsequent sections.
As a bonus stage, select and plot only zones that are close to the centre of London (see Figure 5). Programming encourages rigorous thinking and it helps to define the problem more specifically:
Challenge: Select all zones whose geographic centroid lies within 10 km of the geographic centroid of inner London.[3]
The code below should help understand the way spatial data work in R.
# Find the centre of the london area
easting_lnd <- coordinates(gCentroid(lnd))[[1]]
northing_lnd <- coordinates(gCentroid(lnd))[[2]]
# arguments to test whether or not a coordinate is east or north of the centre
east <- sapply(coordinates(lnd)[,1], function(x) x > easting_lnd)
north <- sapply(coordinates(lnd)[,2], function(x) x > northing_lnd)
# test if the coordinate is east and north of the centre
lnd$quadrant <- "unknown" # prevent NAs in result
lnd$quadrant[east & north] <- "northeast"Challenge: Based on the the above code as refrence try and find the remaining 3 quadrants and colour them as per Figure 6. Hint - you can use the llgridlines function in order to overlay the long-lat lines. For bonus points try to desolve the quadrants so the map is left with only 4 polygons.
The 4 quadrants of London and dissolved borders. Challenge: recreate a plot that looks like this.
Alongside visualisation and interrogation, a GIS must also be able to create and modify spatial data. R's spatial packages provide a very wide and powerful suite of functionality for processing and creating spatial data.
Reprojecting and joining/clipping are fundamental GIS operations, so in this section we will explore how these operations can be undertaken in R. Firstly We will join non-spatial data to spatial data so it can be mapped. Finally we will cover spatial joins, whereby information from two spatial objects is combined based on spatial location.
R objects can be created by entering the name of the class we want to make. vector and data.frame objects for example, can be created as follows:
vec <- vector(mode = "numeric", length = 3)
df <- data.frame(x = 1:3, y = c(1/2, 2/3, 3/4))We can check the class of these new objects using class():
class(vec)## [1] "numeric"
class(df)## [1] "data.frame"
The same logic applies to spatial data. The input must be a numeric matrix or data.frame:
sp1 <- SpatialPoints(coords = df)We have just created a spatial points object, one of the fundamental data types for spatial data. (The others are lines, polygons and pixels, which can be created by SpatialLines, SpatialPolygons and SpatialPixels, respectively.) Each type of spatial data has a corollary that can accepts non-spatial data, created by adding DataFrame. SpatialPointsDataFrame(), for example, creates points with an associated data.frame. The number of rows in this dataset must equal the number of features in the spatial object, which in the case of sp1 is 3.
class(sp1)## [1] "SpatialPoints"
## attr(,"package")
## [1] "sp"
spdf <- SpatialPointsDataFrame(sp1, data = df)
class(spdf)## [1] "SpatialPointsDataFrame"
## attr(,"package")
## [1] "sp"
The above code extends the pre-existing object sp1 by adding data from df. To see how strict spatial classes are, try replacing df with mat in the above code: it causes an error. All spatial data classes can be created in a similar way, although SpatialLines and SpatialPolygons are much more complicated (Bivand et al. 2013). More frequently your spatial data will be read-in from an externally-created file, e.g. using readOGR(). Unlike the spatial objects we created above, most spatial data comes with an associate 'CRS'.
The Coordinate Reference System (CRS) of spatial objects defines where they are placed on the Earth's surface. You may have noticed 'proj4string 'in the summary of lnd above: the information that follows represents its CRS. Spatial data should always have a CRS. If no CRS information is provided, and the correct CRS is known, it can be set as follow:
proj4string(lnd) <- NA_character_ # remove CRS information from lnd
proj4string(lnd) <- CRS("+init=epsg:27700") # assign a new CRSR issues a warning when the CRS is changed. This is so the user knows that they are simply changing the CRS, not reprojecting the data. An easy way to refer to different projections is via EPSG codes.
Under this system 27700 represents the British National Grid. 'WGS84' (epsg:4326) is a very commonly used CRS worldwide. The following code shows how to search the list of available EPSG codes and create a new version of lnd in WGS84:[4]
EPSG <- make_EPSG() # create data frame of available EPSG codes
EPSG[grepl("WGS 84$", EPSG$note), ] # search for WGS 84 code ## code note prj4
## 249 4326 # WGS 84 +proj=longlat +datum=WGS84 +no_defs
## 4890 4978 # WGS 84 +proj=geocent +datum=WGS84 +units=m +no_defs
lnd84 <- spTransform(lnd, CRS("+init=epsg:4326")) # reprojectAbove, spTransform converts the coordinates of lnd into the widely used WGS84 CRS. Now we've transformed lnd into a more widely used CRS, it is worth saving it. R stores data efficiently in .RData or .Rds formats. The former is more restrictive and maintains the object's name, so we use the latter.
# Save lnd84 object (we will use it in Part IV)
saveRDS(object = lnd84, file = "data/lnd84.Rds")Now we can remove the lnd84 object with the rm command. It will be useful later. (In RStudio, notice it also disappears from the Environment in the top right panel.)
rm(lnd84) # remove the lnd object
# we will load it back in later with readRDS(file = "data/lnd84.Rds")Attribute joins are used to link additional pieces of information to our polygons. In the lnd object, for example, we have 4 attribute variables --- that can be found by typing names(lnd). But what happens when we want to add more variables from an external source? We will use the example of recorded crimes by London boroughs to demonstrate this.
To reaffirm our starting point, let's re-load the "london_sport" shapefile as a new object and plot it:
library(rgdal) # ensure rgdal is loaded
# Create new object called "lnd" from "london_sport" shapefile
lnd <- readOGR("data/london_sport.shp")
plot(lnd) # plot the lnd object (not shown)
nrow(lnd) # return the number of rows (not shown)The non-spatial data we are going to join to the lnd object contains records of crimes in London. This is stored in a comma separated values (.csv) file called "mps-recordedcrime-borough". If you open the file in a separate spreadsheet application first, we can see each row represents a single reported crime. We are going to use a function called aggregate to aggregate the crimes at the borough level, ready to join to our spatial lnd dataset. A new object called crime_data is created to store this data.
# Create and look at new crime_data object
crime_data <- read.csv("data/mps-recordedcrime-borough.csv",
stringsAsFactors = FALSE)
head(crime_data$CrimeType) # information about crime type
# Extract "Theft & Handling" crimes and save
crime_theft <- crime_data[crime_data$CrimeType == "Theft & Handling", ]
head(crime_theft, 2) # take a look at the result (replace 2 with 10 to see more rows)
# Calculate the sum of the crime count for each district, save result
crime_ag <- aggregate(CrimeCount ~ Borough, FUN = sum, data = crime_theft)
# Show the first two rows of the aggregated crime data
head(crime_ag, 2)You should not expect to understand all of this upon first try: simply typing the commands and thinking briefly about the outputs is all that is needed at this stage. Here are a few things that you may not have seen before that will likely be useful in the future:
- In the first line of code when we read in the file we specify its location (check in your file browser to be sure).
- The
==function is used to select only those observations that meet a specific condition i.e. where it is equal to, in this case all crimes involving "Theft and Handling". - The
~symbol means "by": we aggregated theCrimeCountvariable by the district name.
Now that we have crime data at the borough level, the challenge is to join it to the lnd object. We will base our join on the Borough variable from the crime_ag object and the name variable from the lnd object. It is not always straight-forward to join objects based on names as the names do not always match. Let's see which names in the crime_ag object match the spatial data object, lnd:
# Compare the name column in lnd to Borough column in crime_ag to see which rows match.
lnd$name %in% crime_ag$Borough## [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [12] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [23] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
# Return rows which do not match
lnd$name[!lnd$name %in% crime_ag$Borough]## [1] City of London
## 33 Levels: Barking and Dagenham Barnet Bexley Brent Bromley ... Westminster
The first line of code above uses the %in% command to identify which values in lnd$name are also contained in the Borough names of the aggregated crime data. The results indicate that all but one of the borough names matches. The second line of code tells us that it is 'City of London'. This does not exist in the crime data. This may be because the City of London has it's own Police Force.[5] (The borough name in the crime data does not match lnd$name is 'NULL'. Check this by typing crime_ag$Borough[!crime_ag$Borough %in% lnd$name].)
Challenge: identify the number of crimes taking place in borough 'NULL', less than 4,000.
Having checked the data found that one borough does not match, we are now ready to join the spatial and non-spatial datasets. It is recommended to use the left_join function from the dplyr package but the merge function could equally be used. Note that when we ask for help for a function that is not loaded, nothing happens, indicating we need to load it:
library(dplyr) # load dplyr We use left_join because we want the length of the data frame to remain unchanged, with variables from new data appended in new columns (see ?left_join). The *join commands (including inner_join and anti_join) assume, by default, that matching variables have the same name. Here we will specify the association between variables in the two data sets:
head(lnd$name) # dataset to add to (results not shown)
head(crime_ag$Borough) # the variables to join
# head(left_join(lnd@data, crime_ag)) # test it works
lnd@data <- left_join(lnd@data, crime_ag, by = c('name' = 'Borough'))## Warning: Column `name`/`Borough` joining factor and character vector,
## coercing into character vector
Take a look at the new lnd@data object. You should see new variables added, meaning the attribute join was successful. Congratulations! You can now plot the rate of theft crimes in London by borough (see Fig 8).
library(tmap) # load tmap package (see Section IV)
qtm(lnd, "CrimeCount") # plot the basic mapNumber of thefts per borough.
Optional challenge: create a map of additional variables in London
With the attribute joining skills you have learned in this section, you should now be able to take datasets from many sources, e.g. data.london.gov.uk, and join them to your geographical data.
In addition to joining by attribute (e.g. Borough name), it is also possible to do spatial joins in R. We use transport infrastructure points as the spatial data to join, with the aim of finding out about how many are found in each London borough.
library(rgdal)
# create new stations object using the "lnd-stns" shapefile.
stations <- readOGR(dsn = "data/lnd-stns.shp")
# stations = read_shape("data/lnd-stns.shp") # from tmap
proj4string(stations) # this is the full geographical detail.
proj4string(lnd) # what's the coordinate reference system (CRS)
bbox(stations) # the extent, 'bounding box' of stations
bbox(lnd) # return the bounding box of the lnd objectThe proj4string() function shows that the Coordinate Reference System (CRS) of stations differs from that of our lnd object. OSGB 1936 (or EPSG 27700) is the official CRS for the UK, so we will convert the 'stations' object to this:
# Create reprojected stations object
stations <- spTransform(stations, CRSobj = CRS(proj4string(lnd)))
plot(lnd) # plot London
points(stations) # overlay the station pointsNote the stations points now overlay the boroughs but that the spatial extent of stations is greater than that of lnd.
To clip the stations so that only those falling within London boroughs are retained we can use sp::over, or simply the square bracket notation for subsetting tabular data (enter ?gIntersects to find out another way to do this):
stations <- stations[lnd, ]
plot(stations) # test the clip succeededgIntersects can achieve the same result, but with more lines of code (see www.rpubs.com/RobinLovelace for more on this) .
tmap was created to overcome some of the limitations of base graphics and ggmap. A concise introduction to tmap can be accessed (after the package is installed) by using the vignette function:
# install.packages("tmap") # install the CRAN version
library(tmap)
vignette("tmap-nutshell")A couple of basic plots show the package's intuitive syntax and attractive default parameters.
qtm(shp = lnd, fill = "Partic_Per", fill.palette = "-Blues") # not shownqtm(shp = lnd, fill = c("Partic_Per", "Pop_2001"), fill.palette = "Blues", ncol = 2) The plot above shows the ease with which tmap can create maps next to each other for different variables. The plot produced by the following code chunk (not shown) demonstrates the power of the tm_facets command. Note that all the maps created with the qtm function can also be created with tm_shape, followed by tm_fill (or another tm_ function).
tm_shape(lnd) +
tm_fill("Pop_2001", thres.poly = 0) +
tm_facets("name", free.coords = TRUE, drop.units = TRUE)To create a basemap with tmap, you can use the read_osm function, from the tmaptools package as follows. Note that you must first transform the data into a geographical CRS:
# Transform the coordinate reference system
lnd_wgs = spTransform(lnd, CRS("+init=epsg:4326"))
if(curl::has_internet()) {
osm_tiles = tmaptools::read_osm(bbox(lnd_wgs)) # download images from OSM
tm_shape(osm_tiles) + tm_raster() +
tm_shape(lnd_wgs) +
tm_fill("Pop_2001", fill.title = "Population, 2001", scale = 0.8, alpha = 0.5) +
tm_layout(legend.position = c(0.89, 0.02))
} else {
tm_shape(lnd_wgs) +
tm_fill("Pop_2001", fill.title = "Population, 2001", scale = 0.8, alpha = 0.5) +
tm_layout(legend.position = c(0.89, 0.02))
}## Warning: Current projection unknown. Long lat coordinates (wgs84) assumed.
Another way to make tmap maps have a basemap is by entering tmap_mode("view"). This will make the maps appear on a zoomable webmap powered by leaflet. There are many other intuitive and powerful functions in tmap. Check the documentation to find out more:
?tmap # get more info on tmapggmap is based on the ggplot2 package, an implementation of the Grammar of Graphics (Wilkinson 2005). ggplot2 can replace the base graphics in R (the functions you have been plotting with so far). It contains default options that match good visualisation practice and is well-documented: http://docs.ggplot2.org/current/ .
As a first attempt with ggplot2 we can create a scatter plot with the attribute data in the lnd object created previously:
library(ggplot2)
p <- ggplot(lnd@data, aes(Partic_Per, Pop_2001))The real power of ggplot2 lies in its ability to add layers to a plot. In this case we can add text to the plot.
p + geom_point(aes(colour = Partic_Per, size = Pop_2001)) +
geom_text(size = 2, aes(label = name))This idea of layers (or geoms) is quite different from the standard plot functions in R, but you will find that each of the functions does a lot of clever stuff to make plotting much easier (see the documentation for a full list).
In the following steps we will create a map to show the percentage of the population in each London Borough who regularly participate in sports activities.
ggmap requires spatial data to be supplied as data.frame, using tidy(). The generic plot() function can use Spatial* objects directly; ggplot2 cannot. Therefore we need to extract them as a data frame. The tidy function was written specifically for this purpose. For this to work, broom package must be installed.
lnd_f <- broom::tidy(lnd)## Regions defined for each Polygons
This step has lost the attribute information associated with the lnd object. We can add it back using the left_join function from the dplyr package (see ?left_join).
head(lnd_f, n = 2) # peak at the fortified data
lnd$id <- row.names(lnd) # allocate an id variable to the sp data
head(lnd@data, n = 2) # final check before join (requires shared variable name)
lnd_f <- left_join(lnd_f, lnd@data) # join the data## Joining, by = "id"
The newlnd_f object contains coordinates alongside the attribute information associated with each London Borough. It is now straightforward to produce a map with ggplot2. coord_equal() is the equivalent of asp = T in regular plots with R:
map <- ggplot(lnd_f, aes(long, lat, group = group, fill = Partic_Per)) +
geom_polygon() + coord_equal() +
labs(x = "Easting (m)", y = "Northing (m)",
fill = "% Sports\nParticipation") +
ggtitle("London Sports Participation")Entering map should result in your first ggplot-made map of London. The default colours are really nice but we may wish to produce the map in black and white, which should produce a map like the one shown below. Try changing the colours and saving plots with ggsave().
map + scale_fill_gradient(low = "white", high = "black")Leaflet is the world's premier web mapping system, serving hundreds of thousands of maps worldwide each day. The JavaScript library actively developed at github.com/Leaflet/Leaflet, has a strong user community. It is fast, powerful and easy to learn.
The leaflet package creates interactive web maps in few lines of code. One of the exciting things about the package is its tight integration with the R package for interactive on-line visualisation, shiny. Used together, these allow R to act as a complete map-serving platform, to compete with the likes of GeoServer! For more information on rstudio/leaflet, see rstudio.github.io/leaflet/ and the following on-line tutorial: robinlovelace.net/r/2015/02/01/leaflet-r-package.html.
install.packages("leaflet")
library(leaflet)
lnd84 <- readRDS('data/lnd84.Rds')
leaflet() %>%
addTiles() %>%
addPolygons(data = lnd84)The below code demonstrates how to read in the necessary data for this task and 'tidy' it up. The data file contains historic population values between 1801 and 2001 for London, again from the London data store.
We tidy the data so that the columns become rows. In other words, we convert the data from 'flat' to 'long' format, which is the form required by ggplot2 for faceting graphics: the date of the population survey becomes a variable in its own right, rather than being strung-out over many columns.
london_data <- read.csv("data/census-historic-population-borough.csv")
# install.packages("tidyr")
library(tidyr) # if not install it, or skip the next two steps
ltidy <- gather(london_data, date, pop, -Area.Code, -Area.Name)
head(ltidy, 2) # check the output (not shown)In the above code we take the london_data object and create the column names 'date' (the date of the record, previously spread over many columns) and 'pop' (the population which varies). The minus (-) symbol in this context tells gather not to include the Area.Name and Area.Code as columns to be removed. In other words, "leave these columns be". Data tidying is an important subject: more can be read on the subject in Wickham (2014) or in a vignette about the package, accessed from within R by entering vignette("tidy-data").
Merge the population data with the London borough geometry contained within our lnd_f object, using the left_join function from the dplyr package:
head(lnd_f, 2) # identify shared variables with ltidy ## long lat order hole piece group id ons_label name
## 1 541177.7 173555.7 1 FALSE 1 0.1 0 00AF Bromley
## 2 541872.2 173305.8 2 FALSE 1 0.1 0 00AF Bromley
## Partic_Per Pop_2001 quadrant CrimeCount
## 1 21.7 295535 southeast 15172
## 2 21.7 295535 southeast 15172
ltidy <- rename(ltidy, ons_label = Area.Code) # rename Area.code variable
lnd_f <- left_join(lnd_f, ltidy)## Joining, by = "ons_label"
## Warning: Column `ons_label` joining factors with different levels, coercing
## to character vector
Rename the date variable (use ?gsub and Google 'regex' to find out more).
lnd_f$date <- gsub(pattern = "Pop_", replacement = "", lnd_f$date)We can now use faceting to produce one map per year:
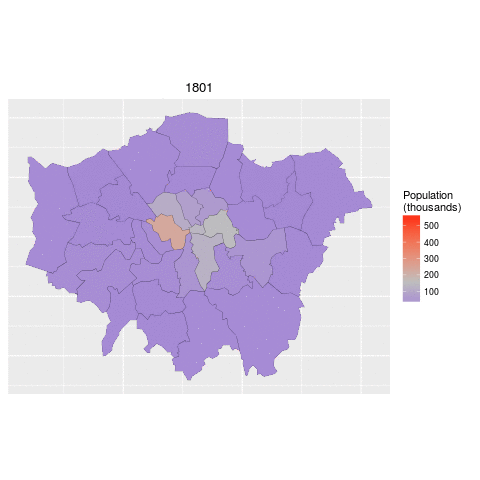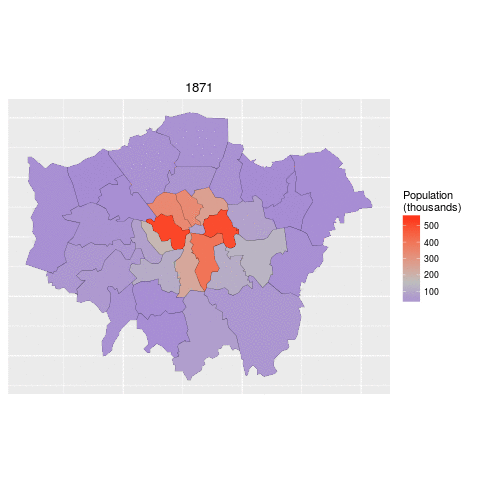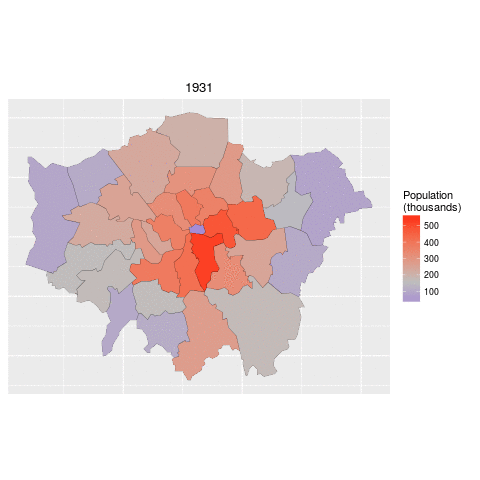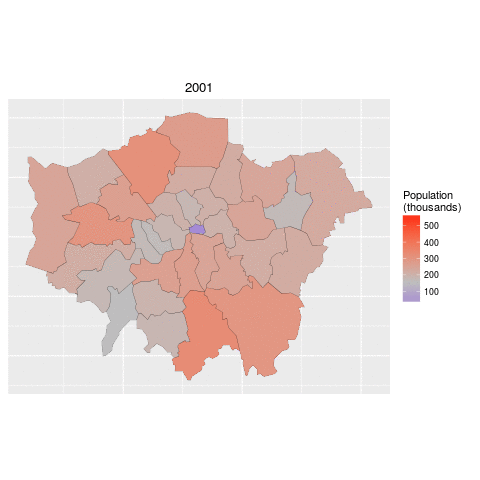
ggplot(data = lnd_f, # the input data
aes(x = long, y = lat, fill = pop/1000, group = group)) + # define variables
geom_polygon() + # plot the boroughs
geom_path(colour="black", lwd=0.05) + # borough borders
coord_equal() + # fixed x and y scales
facet_wrap(~ date) + # one plot per time slice
scale_fill_gradient2(low = "blue", mid = "grey", high = "red", # colors
midpoint = 150, name = "Population\n(thousands)") + # legend options
theme(axis.text = element_blank(), # change the theme options
axis.title = element_blank(), # remove axis titles
axis.ticks = element_blank()) # remove axis ticks# ggsave("figure/facet_london.png", width = 9, height = 9) # save figureThere is a lot going on here so explore the documentation to make sure you understand it. Try out different colour values as well.
Try experimenting with the above code block to see what effects you can produce.
Challenge 1: Try creating this plot for the % of population instead of the absolute population.
Challenge 2: For bonus points, try creating an animation of London's evolving population over time (hint: a file called ggnaminate.R may help).
The skills taught in this tutorial are applicable to a very wide range of situations, spatial or not. Often experimentation is the most rewarding learning method, rather than just searching for the 'best' way of doing something (Kabakoff, 2011). We recommend you play around with your data.
If you enjoyed this tutorial, you may find the book chapter "Spatial Data Visualisation with R" of interest (Cheshire and Lovelace, 2014). The project's repository can be found on its GitHub page: github.com/geocomPP/sdvwR. There are also a number of bonus 'vignettes' associated with the present tutorial. These can be found on the vignettes page of the project's repository.
Other advanced tutorials include
- The Simple Features vignettes.
- "solaR: Solar Radiation and Photovoltaic Systems with R", a technical academic paper on the solaR package which contains a number of spatial functions.
Such tutorials are worth doing as they will help you understand R's spatial 'ecosystem' as a cohesive whole rather than as a collection of isolated functions. In R the whole is greater than the sum of its parts.
The supportive on-line communities surrounding large open source programs such as R are one of their greatest assets, so we recommend you become an active "open source citizen" rather than a passive consumer.
Good resources that will help you further sharpen you R skills include:
-
R's homepage hosts a wealth of official and contributed guides. http://cran.r-project.org
-
StackOverflow and GIS.StackExchange groups (the "[R]" search term limits the results). If your question has not been answered yet, just ask, preferably with a reproducible example.
-
R's mailing lists, especially R-sig-geo. See r-project.org/mail.html.
-
Dorman (2014): detailed exposition of spatial data in R, with a focus on raster data. A free sample of this book is available online.
-
Bivand et al. (2013) : 'Applied spatial data analysis with R' - provides a dense and detailed overview of spatial data analysis.
The tutorial was developed for a series of Short Courses funded by the National Centre for Research Methods (NCRM), via the TALISMAN node (see geotalisman.org). Thanks to the ESRC for funding applied methods research. Many thanks to Matt Whittle, Alistair Leak, Hannah Roberts and Phil Jones who helped develop and demonstrate these materials. Amy O'Neill organised the course and encouraged feedback from participants. The final thanks is to all users and developers of open source software for making powerful tools such as R accessible and enjoyable to use.
If you have found this tutorial useful in your work, please cite it:
Lovelace, R., & Cheshire, J. (2014). Introduction to visualising spatial data in R. National Centre for Research Methods Working Papers, 14(03). Retrieved from https://github.com/Robinlovelace/Creating-maps-in-R
Bivand, R. S., Pebesma, E. J., & Rubio, V. G. (2013). Applied spatial data analysis with R. Springer. 2nd ed.
Cheshire, J., & Lovelace, R. (2015). Spatial data visualisation with R. In C. Brunsdon & A. Singleton (Eds.), Geocomputation (pp. 1–14). SAGE Publications. Retrieved from https://github.com/geocomPP/sdv . Full chapter available from https://www.researchgate.net/publication/274697165_Spatial_data_visualisation_with_R
Dorman, M. (2014). Learning R for Geospatial Analysis. Packt Publishing Ltd.
Gillespie, Colin, and Robin Lovelace. 2016. Efficient R Programming: A Practical Guide to Smarter Programming. O’Reilly Media. https://csgillespie.github.io/efficientR/.
Kabacoff, R. (2011). R in Action. Manning Publications Co.
Lamigueiro, O. P. (2012). solaR: Solar Radiation and Photovoltaic Systems with R. Journal of Statistical Software, 50(9), 1–32. Retrieved from http://www.jstatsoft.org/v50/i09
Wickham, H. (2014). Tidy data. The Journal of Statistical Software, 14(5), Retrieved from http://www.jstatsoft.org/v59/i10
Wilkinson, L. (2005). The grammar of graphics. Springer.
[1] Tip: typing Alt - on the keyboard will create the arrow in RStudio. The equals sign = also works.
[2] The third line of code is included for historical interest and to provide an opportunity to discuss R functions and their arguments (the values inside a function's brackets) in detail. Note the arguments are separated by a comma. The order in which they are specified is important. You do not have to explicitly type dsn = or layer = as R knows which order they appear. readOGR("data", "london_sport") would work just as well. For clarity, it is good practice to include argument names when learning new functions so we will continue to do so.
[3] To see how this map was created, see the code in README.Rmd . This may be loaded by typing file.edit("README.Rmd") or online at github.com/Robinlovelace/Creating-maps-in-R/blob/master/README.Rmd.
[4] Note: entering projInfo() provides additional CRS options. spatialreference.org provides more information about EPSG codes.
[5] See www.cityoflondon.police.uk/.