-
-
Notifications
You must be signed in to change notification settings - Fork 208
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
1 parent
8008f3a
commit 3d11a70
Showing
8 changed files
with
181 additions
and
36 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,22 @@ | ||
| # `BayesianPINN` Discretizer for PDESystems | ||
|
|
||
| Using the Bayesian PINNs solvers, we can solve general nonlinear PDEs,ODEs and Also simultaniously perform PDE,ODE parameter Estimation. | ||
|
|
||
| Note: The BPINN PDE solver also works for ODEs defined using ModelingToolkit, [ModelingToolkit.jl PDESystem documentation](https://docs.sciml.ai/ModelingToolkit/stable/systems/PDESystem/). Despite this the ODE specific BPINN solver `BNNODE` [refer](https://docs.sciml.ai/NeuralPDE/dev/manual/ode/#NeuralPDE.BNNODE) exists and uses `NeuralPDE.advancedhmc_pinn_ode` at a lower level. | ||
|
|
||
| # `BayesianPINN` Discretizer for PDESystems and lower level Bayesian PINN Solver calls for PDEs and ODEs. | ||
|
|
||
| ```@docs | ||
| NeuralPDE.BayesianPINN | ||
| NeuralPDE.advancedhmc_pinn_pde | ||
| NeuralPDE.advancedhmc_pinn_ode | ||
| ``` | ||
|
|
||
| ## `symbolic_discretize` for `BayesianPINN` and lower level interface. | ||
|
|
||
| ```@docs | ||
| SciMLBase.symbolic_discretize(::PDESystem, ::NeuralPDE.AbstractPINN) | ||
| NeuralPDE.BPINNstats | ||
| NeuralPDE.BPINNsolution | ||
| ``` | ||
|
|
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
2 changes: 1 addition & 1 deletion
2
docs/src/tutorials/low_level.md → docs/src/tutorials/low_level_1.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,75 @@ | ||
| # Using `ahmc_bayesian_pinn_pde` with the `BayesianPINN` Discretizer for the 1-D Burgers' Equation | ||
|
|
||
| Let's consider the Burgers' equation: | ||
|
|
||
| ```math | ||
| \begin{gather*} | ||
| ∂_t u + u ∂_x u - (0.01 / \pi) ∂_x^2 u = 0 \, , \quad x \in [-1, 1], t \in [0, 1] \, , \\ | ||
| u(0, x) = - \sin(\pi x) \, , \\ | ||
| u(t, -1) = u(t, 1) = 0 \, , | ||
| \end{gather*} | ||
| ``` | ||
|
|
||
| with Bayesian Physics-Informed Neural Networks. Here is an example of using `BayesianPINN` discretization with `ahmc_bayesian_pinn_pde` : | ||
|
|
||
| ```@example low_level_2 | ||
| using NeuralPDE, Lux, ModelingToolkit | ||
| import ModelingToolkit: Interval, infimum, supremum | ||
| @parameters t, x | ||
| @variables u(..) | ||
| Dt = Differential(t) | ||
| Dx = Differential(x) | ||
| Dxx = Differential(x)^2 | ||
| #2D PDE | ||
| eq = Dt(u(t, x)) + u(t, x) * Dx(u(t, x)) - (0.01 / pi) * Dxx(u(t, x)) ~ 0 | ||
| # Initial and boundary conditions | ||
| bcs = [u(0, x) ~ -sin(pi * x), | ||
| u(t, -1) ~ 0.0, | ||
| u(t, 1) ~ 0.0, | ||
| u(t, -1) ~ u(t, 1)] | ||
| # Space and time domains | ||
| domains = [t ∈ Interval(0.0, 1.0), | ||
| x ∈ Interval(-1.0, 1.0)] | ||
| # Discretization | ||
| dx = 0.05 | ||
| # Neural network | ||
| chain = Lux.Chain(Lux.Dense(2, 10, Lux.σ), Lux.Dense(10, 10, Lux.σ), Lux.Dense(10, 1)) | ||
| strategy = NeuralPDE.GridTraining([dx,dx]) | ||
| discretization = NeuralPDE.BayesianPINN([chain], strategy) | ||
| @named pde_system = PDESystem(eq, bcs, domains, [x, t], [u(x, t)]) | ||
| sol1 = ahmc_bayesian_pinn_pde(pde_system, | ||
| discretization; | ||
| draw_samples = 100, | ||
| bcstd = [0.01, 0.03, 0.03, 0.01], | ||
| phystd = [0.01], | ||
| priorsNNw = (0.0, 10.0), | ||
| saveats = [1 / 100.0, 1 / 100.0],progress=true) | ||
| ``` | ||
|
|
||
| And some analysis: | ||
|
|
||
| ```@example low_level | ||
| using Plots | ||
| ts, xs = [infimum(d.domain):0.01:supremum(d.domain) for d in domains] | ||
| u_predict_contourf = reshape([first(phi([t, x], res.u)) for t in ts for x in xs], | ||
| length(xs), length(ts)) | ||
| plot(ts, xs, u_predict_contourf, linetype = :contourf, title = "predict") | ||
| u_predict = [[first(phi([t, x], res.u)) for x in xs] for t in ts] | ||
| p1 = plot(xs, u_predict[3], title = "t = 0.1"); | ||
| p2 = plot(xs, u_predict[11], title = "t = 0.5"); | ||
| p3 = plot(xs, u_predict[end], title = "t = 1"); | ||
| plot(p1, p2, p3) | ||
| ``` | ||
|
|
||
| 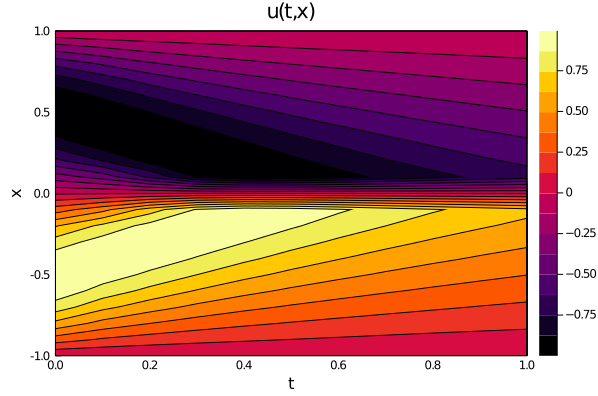 | ||
|
|
||
| 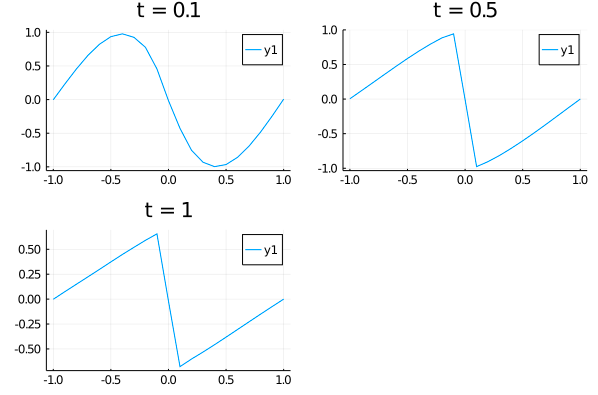 |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters