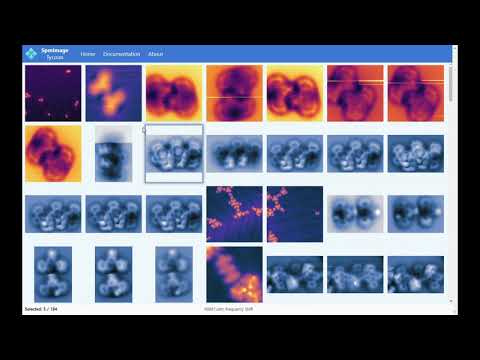
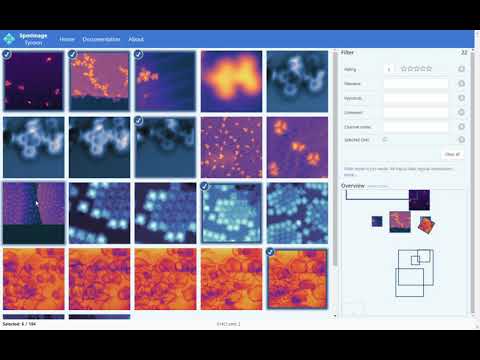
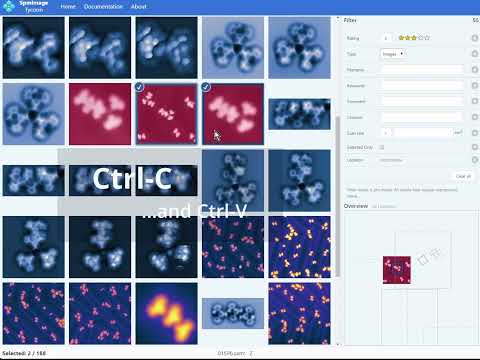
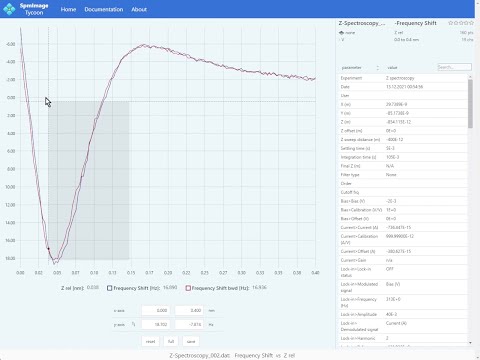
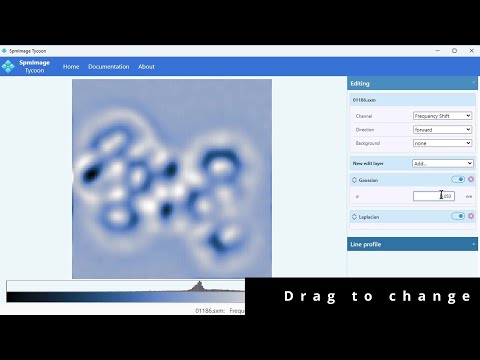
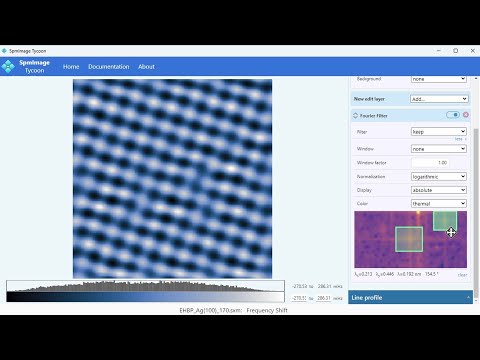
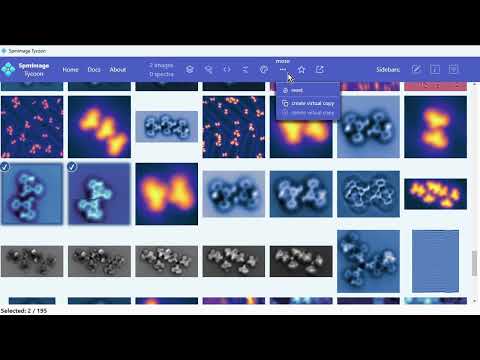
Cross-platform app to manage and edit scanning probe microscopy (SPM) images and spectra. It supports scanning tunneling microscopy (STM) and atomic force microscopy (AFM) images and spectra.
Features
Demo
Supported file formats
Disclaimer
Installation
Installation (alternative)
Cite
Case study
Tips and tricks
Accessing the database
Technical background
Third party libraries
Contribute
Related projects
- Load and display SPM images: grid and zoom views
- Load and display spectra
- Fast navigation: dropdown menus or keyboard shortcuts
- Cycle through channels, switch between forward and backward directions
- Different types of background corrections
- Various color palettes
- Non-destructive edits: Gaussian, Laplacian, Fourier Filter, etc.
- Line profiles
- Star-rating and keyword systems
- Powerful search and filtering (e.g., by keywords, data type, location, scan size)
- Overview and position-based filtering
- Export to OpenDocument presentation (compatible with LibreOffice, OpenOffice, PowerPoint, etc.)
- Everything is experimental. And we all like experiments.
It is best to play around with the app yourself, so go ahead and install it.. Example data can be found in the test directory.
| images | spectra | ||
| Nanonis | .sxm | .dat | release |
| GXSM | .nc | .vpdata | testing |
| Asylum Research | .ibw | testing |
Currently, Nanonis files have the most extensive support. GXSM and Asylum Research file support is implemented and needs further testing. If you want to contribute to the development or testing, please get in touch with me. If you come across any issues or have suggestions for other file formats, please let me know.
Currently, the app should be considered experimental. It has undergone limited testing and I can not guarantee that it will work flawlessly. It is conceivable that some of the calculations potentially give erroneous results under certain circumstances.
The app never modifies or deletes your original data. However, in case of any unexpected problems, you might lose the modifications saved within the app. Even though this has never happened to me, I still advice to backup the database regularly (I personally do not do that, though). The app creates a database in each project directory under _spmimages_cache/db.jld2 (some older versions of this file are kept as well). This file contains all your edits, keywords, etc. and can be copied as a backup. Any filesystem backup solution should handle this.
Please only use the app if you read the disclaimer above and feel brave enough to do so.
Use SpmImage Tycoon Installer. It will install a compiled version of SpmImage Tycoon with much improved startup speed. Furthermore, it can create startmenu and desktop shortcuts. This way of installation also allows to copy the app from one computer to another, by simply copying the installation directory.
Usage is simple:
- Install Julia
- Start Julia and type the following two commands:
using Pkg
Pkg.add("SpmImageTycoonInstaller")
using SpmImageTycoonInstaller
install()This will install a compiled version of SpmImage Tycoon. The installation will typically take 10 to 20 minutes and take up around 1 GB of space. The same procedure can be used to update to the latest version.
As an alternative to the automatic installation process described above, you can manually install SpmImage Tycoon:
- Install Julia
- Start Julia and type the following two commands:
using Pkg
Pkg.add("SpmImageTycoon")- To run the app, type (within Julia):
using SpmImageTycoon
tycoon()(if there are any firewall requests, please accept them - it is only for local server/client communication)
- As a more convenient alternative to 3, just use one of the scripts in the helpers directory to start the app. There is a shell script for linux, as well as a bat script for windows and also an autohotkey script. You can find more information on autohotkey here.
- Start Julia and type the following two commands:
using Pkg
Pkg.update()- done
If you use the app for your scientific work, please consider citing it:
@article{Riss_JOSS_2022,
doi = {10.21105/joss.04644},
url = {https://doi.org/10.21105/joss.04644},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {77},
pages = {4644},
author = {Alexander Riss},
title = {SpmImage Tycoon: Organize and analyze scanning probe microscopy data},
journal = {Journal of Open Source Software}
}- Fast access to the file system is crucial - do not use this app on a slow networked file system.
- The first startup after installation (or update) can be slow - it is due to initial compilations. I am working on a solution to this.
- To gain a significant speedup through mutlithreading, start Julia with multiple threads, e.g.:
julia --threads=auto. This is automatically done by the SpmImage Tycoon Installer. - There is a small overhead to apply color schemes; for performance it is best to use the standard "gray" color scheme.
- Extra settings (such as default channels and the default color scheme) can be changed in the
<homedir>/.spmimagetycoon/settings.tomlfile.
The app uses a HDF5-compatible database, which can be read using Python, Julia, or any other language that supports HDF5. An example of how to access the database using Python is shown here.
The app is written in Julia, using Blink.jl for the Electron-based frontend.
The Julia source code is in the src directory, the HTML, CSS, and JavaScript source code for the frontend is in the res directory.
The program relies on
- SpmImages.jl: Julia library to read and display SPM images.
- SpmSpectroscopy.jl: Julia library to read and analyze SPM spectra.
The following javascript and css libraries are included in the app:
- Bulma: MIT License
- μPlot: MIT License
- SelectionJS: MIT License
- Simple-DataTables: LGPL License
- Sortable: MIT License
- tagify: MIT License
Contributions are very welcome. Please post issues, suggestions, and pull requests on github. If you want to contribute to the code, take a look at the technical background and contribution guidelines; and please get in touch with me.
Follow me on X (twitter) for updates and more information about this project:
- SpmImages.jl: Julia library to read and display SPM images.
- SpmSpectroscopy.jl: Julia library to read and analyze SPM spectra.
- SpmGrids.jl: Julia library to read and analyze SPM grid spectroscopy.
- imagex: Python scripts to analyze scanning probe images.
- gridex: Python scripts to analyze 3D grid data.