-
Arthur Alexandro Soenarto
-
Gabriel El Haddad
-
Xiaoning Wang
-
Syed Taha Ali
We choose the HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. It is also the dataset of the International Skin Imaging Collaboration (ISIC) 2018 challenge.
cd into folder then,
$ virtualenv -p `which python3.8` venv/
$ source venv/bin/activate
$ pip install -r requirements.txt
$ deactivate # when done $ ssh [email protected]
$ virtualenv -p `which python3.8` venv/
$ source venv/bin/activate
$ pip install -r requirements.txt
$ deactivate
$ sbatch segtrainjob.sh # modify segtrainjob.sh with file u want to run
$ squeue --user=csc490w -t RUNNING # status of running jobIf some of the libraries do not install correctly, need to download them using their Compute Canada alias
$ avail_wheels "*name*" # some libraries have different versions for cpu & gpu
$ pip install <name> --no-indexunder main() at end of file:
- change the training images and ground truth file path based on directory structure
- change batch_sizes and iter_sizes based on what you want to train on
- change file path and name of where you want to store your pretrained models
at the end of def train():
- under if plot:, change file path and name of where you want to store your loss curve
under main() at end of file:
- change the training images and ground truth file path based on directory structure
- change the validation images and ground truth file path based on directory structure
- change doubleunet_models and model.load_state_dict() in the for loop to point to where the pretrained doubleunet models are based on directury structure
- change unet_model_path to point to where the pretrained singleunet are based on directury structure
- under f = open(...) before the for loop: change file path and name to where you want to store your analysis.
optional:
- you can skip step no.1 because training takes too long
To run the code, follow the python notebook to train the model. Follow the python notebook to get the result. To try different models, you can simply replace the model variable to see the result for different models. For CapsFix mode, it is recommanded to thier origional github link to test out the results: (https://github.com/Woodman718/FixCaps)
This project is an application of different machine learning models that were used for attempting the 2018 ISIC Challenge.
This challenge is broken into three separate tasks:
- Task 1: Lesion Segmentation
- Task 2: Lesion Attribute Detection
- Task 3: Disease Classification
We decided to combine the tasks where the final goal was classification (task 3).
Skin Cancer is one of the most common cancers in North America. The most common cause is from overexposure to ultraviolet rays from the sun. It involves the growth of abnormal cells in the outermost skin layer (called the epidermis), which can form malignant tumors if not treated early. Since it grows in the outermost layer, this property makes skin cancer easily detectable and extremely relevant to camera based machine learning applications, which is the motivation for our project.
Sequence diagram:
Segmentation is the process of associating each pixel on an image with a class label, For task 1, our target label was the image mask of the skin lesion, so we had to perform binary segmentation.
The input data are dermoscopic lesion images in JPEG format. All lesion images are named using the scheme ISIC_<image_id>.jpg. The response data are binary mask images in PNG format. Mask images are named using the scheme ISIC_<image_id>_segmentation.png, where <image_id> matches the corresponding lesion image for the mask.
- 0: representing the background of the image, or areas outside the primary lesion
- 255: representing the foreground of the image, or areas inside the primary lesion
For training, the image was resized to 90x90 with various transformations applied and normalized such that (0, 255) -> (0, 1).
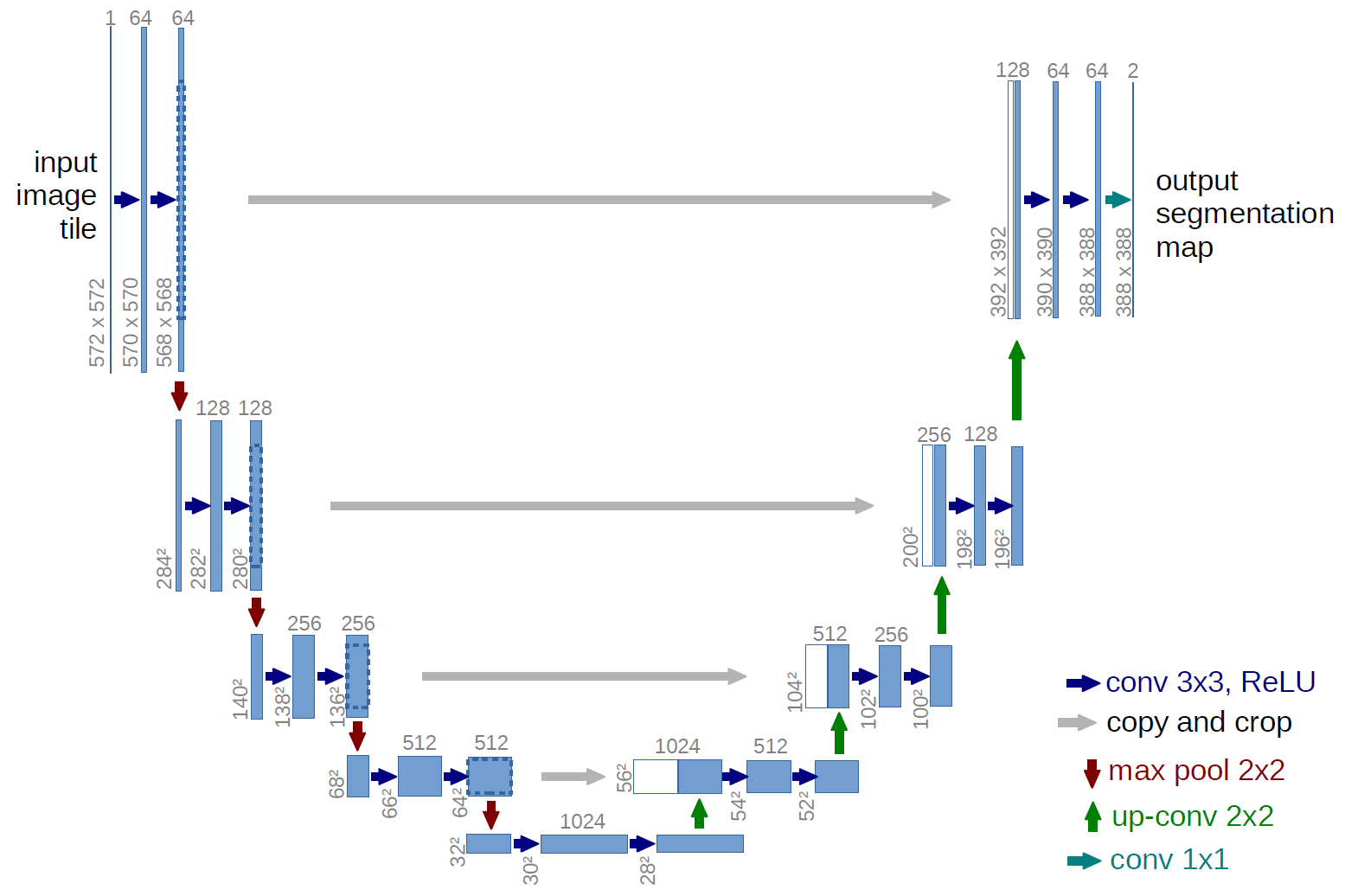
We first implemented UNet, a convolutional network architecture for fast and precise segmentation of images. U-Net was developed for biomedical image segmentation at the Computer Science Department of the University of Freiburg. The network is a convolutional neural network.
Model:
Then, we explored a more advanced version of UNet, the DoubleUNet, which is a combination of two U-Net architectures stacked on top of each other.
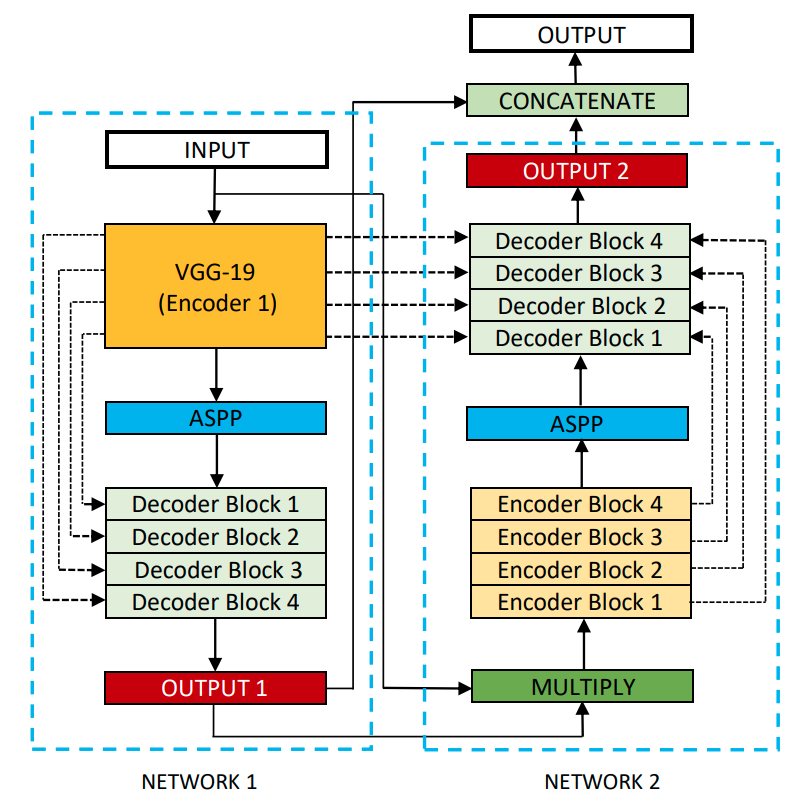 source: https://raw.githubusercontent.com/DebeshJha/2020-CBMS-DoubleU-Net/master/img/DoubleU-Net.png
source: https://raw.githubusercontent.com/DebeshJha/2020-CBMS-DoubleU-Net/master/img/DoubleU-Net.png
Finally, we also tried a novel approach of a Triple UNet by combining the output of the single UNet and Double UNet.
Since the output images pixels are either 0's (if part of the background) or 1's (if part of the lesion), the two combination approaches we tested:
- Intersection of the predicted lesion regions of the two models using logical and
- Union of the prediction lesion regions using logical or
We had greater validation accuracy with the union of the two masks, so we decided to use union for the Triple UNet.
| Segmentation Model | Accuracy | Dice Score | IOU |
|---|---|---|---|
| UNet (batch=100, epoch=500) | 0.8691 | 0.7385 | 0.6189 |
| Double UNet (batch=64, epoch=50) | 0.8590 | 0.7194 | 0.6020 |
| Triple UNet (union) | 0.8712 | 0.7433 | 0.6317 |
Similarly to how physicians provided a ground truth mask dataset for our general segmentation training, the isic 2018 dataset also provides ground truth masks for specific attributes.
The following dermoscopic attributes can be identified:
- pigment network
- negative network
- streaks
- milia-like cysts
- globules (including dots)
The input data are dermoscopic lesion images in JPEG format. All lesion images are named using the scheme ISIC_<image_id>.jpg, where <image_id>. The response data are binary mask images in PNG format, indicating the location of a dermoscopic attribute within each input lesion image. Mask images are named using the scheme ISIC_<image_id>attribute<attribute_name>.png, where <image_id> matches the corresponding lesion image for the mask and <attribute_name> identifies a dermoscopic attribute (pigment_network, negative_network, streaks, milia_like_cyst, and globules).
Similarly to the segmentation data, the mask image ground truth pixels are either 0 (indicating areas where attribute is absent) and 255 (where the attribute is present), hence similar transformations to the segmentation task were applied here as well.
This is a segmentation problem again, so a UNet was selected again to detect the attributes.
| Mask Type | Number of Blank Images | % Blank Images | Pixel Ratio |
|---|---|---|---|
| Pigment network | 1992 | 77 | 0.0309 |
| Negative network | 1915 | 74 | 0.01272 |
| Streaks | 2405 | 93 | 0.0067 |
| Milia like cyst | 1072 | 41 | 0.1370 |
| Globules | 2494 | 96 | 0.0042 |
Dataset is severely imbalanced with a lot of blank images and even in the images where they do appear, the relative sizes of some attributes are small. We decided not to include attributes as a feature for the final task.
The attribute segmentation would have been used alongside the overall segmentation like so:

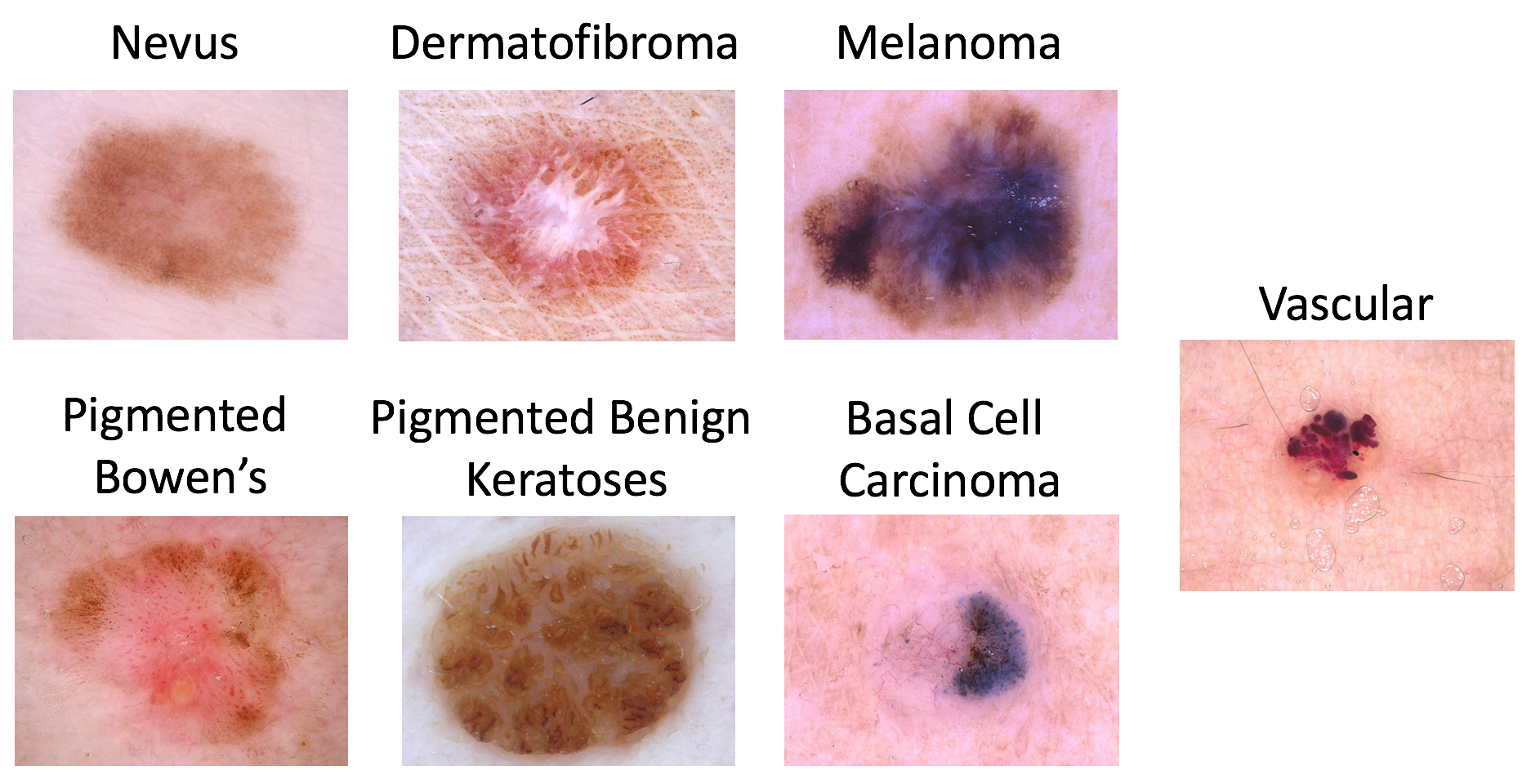
There are 7 possible diagnosis categories for a skin lesion sample:
- Melanoma
- Melanocytic nevus
- Basal cell carcinoma
- Actinic keratosis / Bowens disease (intraepithelial carcinoma)
- Benign keratosis (solar lentigo / seborrheic keratosis / lichen planus-like keratosis)
- Dermatofibroma
- Vascular lesion
The training data is provided as a csv, where each row is a sample containing the attributes as columns:
- image: an input image identifier of the form ISIC_
- MEL: "Melanoma" diagnosis confidence
- NV: "Melanocytic nevus" diagnosis confidence
- BCC: "Basal cell carcinoma" diagnosis confidence
- AKIEC: "Actinic keratosis / Bowen's disease (intraepithelial carcinoma)" diagnosis confidence
- BKL: "Benign keratosis (solar lentigo / seborrheic keratosis / lichen planus-like keratosis)" diagnosis confidence
- DF: "Dermatofibroma" diagnosis confidence
- VASC: "Vascular lesion" diagnosis confidence
For this task, the segmentation model was first used to produce a predicted mask for the image, which was then overlayed onto the image.
The overlay was applied by multiplying the input image and mask (similar to logical and), where the background region that has a pixel value of 0 when multiplied with the input image would yield 0 and the lesion region that has a pixel value of 1 when multiplied with the input image would yield the input images original pixel value.
Then the classification model was trained on the transformed image.
As our final task of our work, we tried out different models to do the classification work. We tried different types of models to learn the dataset robustly.
We first implemented ResNet, a residual network that works very well on classification tasks. However, convolutional networks also have significant drawbacks. Capsule networks are one of the methods to compensate for the shortcomings of CNNs. The critical difference is that there are no max-pooling layers in the network. Instead, the capsule network will have unique components called Capsules. Each capsule represent is a vector that represents the a label and we can use three dense layer to reconstruct the image.
For the best accuracy, we used the FixCap that has the highest accuracy for the dataset. (https://github.com/Woodman718/FixCaps). They used an optimized capsule network:
We also tried with different methods to improve the ResNet for example, replace the regular convolution operation to deformable convolution. The intuition here is that the deformable will have offsets to let the convolution to concentrate on the disease instead of other part of skins.
The final accuray is shown as below, and it turns out our approch is not successful to improve the accuracy. The future work would be validating the novel DeformCaps Network and try in simpler datasets.
Arthur - Implemented and trained DoubleUNet, TripleUNet and organized the poster template
Taha - Implemented and trained UNet, data augmentation, gathered metrics
Gabriel - Attribute detection, data visualization
Xiaoning - Implemented classification models
- Skin Cancer - Incidence Rates (2022) https://www.aad.org/media/stats-skin-cancer
- Das. K et al., Machine Learning and Its Application in Skin Cancer (2021) www.ncbi.nlm.nih.gov/pmc/articles/PMC8705277
- Tschandl, P., Rosendahl, C. & Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. (2018).
- O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” (MICCAI), 2015
- Jha. Debesh, et. al. DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation (2020)
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep Residual Learning for Image Recognition. 2016 IEEE CONFERENCE ON COMPUTER VISION AND PATTERN RECOGNITION (CVPR), 770–778. https://doi.org/10.1109/CVPR.2016.90
- Sabour, S., Frosst, N., & Hinton, G. E. (2017). Dynamic Routing Between Capsules.
- Z. Lan, S. Cai, X. He and X. Wen, "FixCaps: An Improved Capsules Network for Diagnosis of Skin Cancer," in IEEE Access, vol. 10, pp. 76261-76267, 2022, doi: 10.1109/ACCESS.2022.3181225.
- Li, C., Zhang, D., Tian, Z., Du, S., & Qu, Y. (2020). Few‐shot learning with deformable convolution for multiscale lesion detection in mammography. Medical Physics (Lancaster), 47(7), 2970–2985. https://doi.org/10.1002/mp.14129