-
Notifications
You must be signed in to change notification settings - Fork 7
TreeSlicer: Example 2
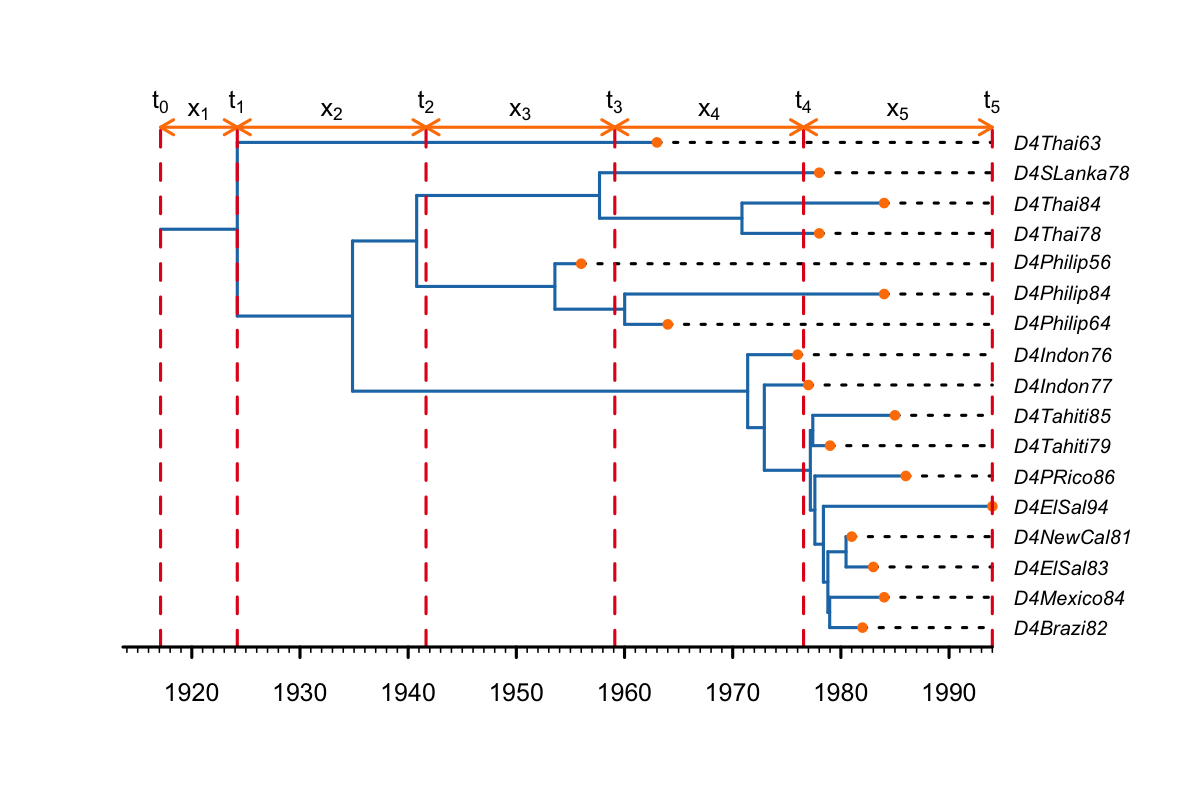
In this example we set change-point times to be equally spaced between the tMRCA and the most recent sample (the present, as in the figure below. This is usually used for the effective reproductive number (Re) or the birth-rate. Thus, shifts in the Re are placed at equidistant intervals between the tMRCA and the present (t1-t4), i.e. the time between the tMRCA (1924) and t1 (1938) is the same as the time between each of the other shifts (e.g. t2 and t3, 1952-1966). Thus, only the first interval (between t0 and t1) is longer than the others, because it incorporates the origin branch and also includes the tMRCA.
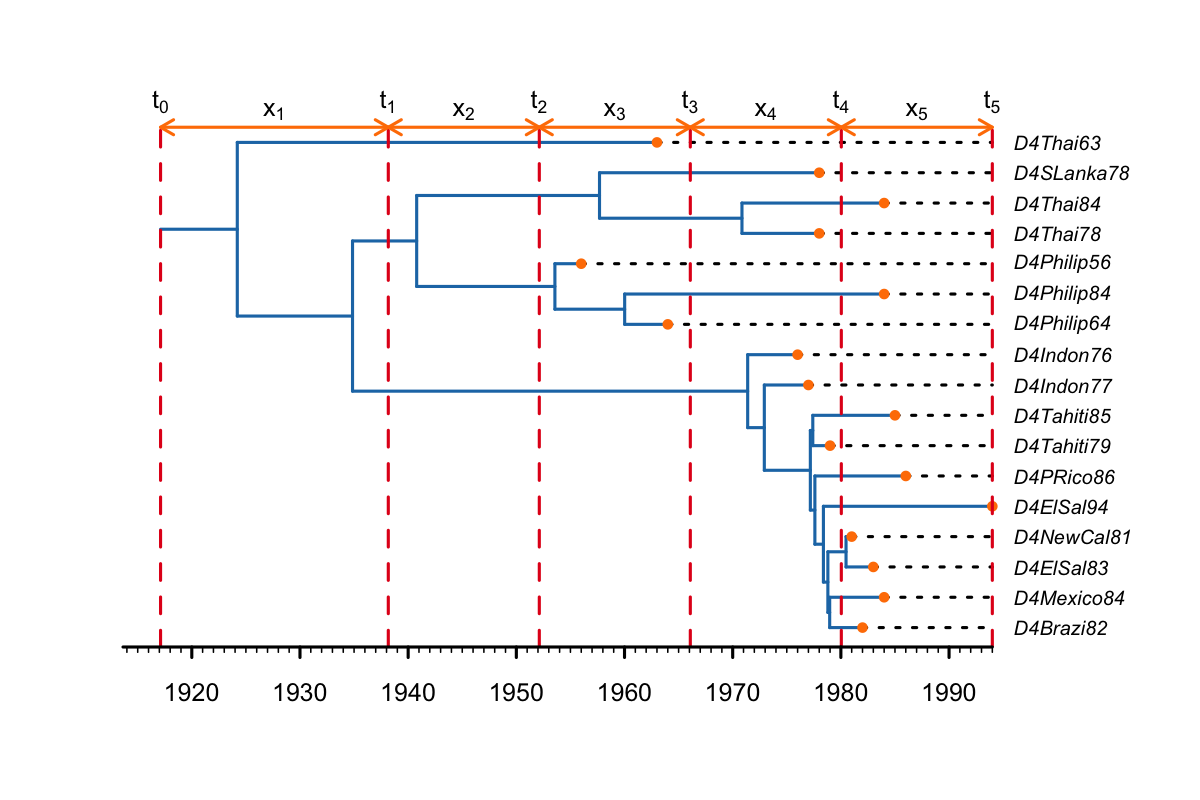
Keep in mind that the time of the origin represents the start of the outbreak, which is always bigger than the tMRCA of the sampled tree. If all infections are in the sampled tree (i.e. the sampled tree is the complete tree), the tMRCA would represent the time of the first transmission from the index case to cause the second infection. The origin represents the time the index case became infected. (In methods based on the Kingman coalescent the origin is not estimated and the process stops at the tMRCA of the sampled tree).
The default behaviour of BDSKY is to place change-point times at equally spaced intervals between the origin and the present, as below.
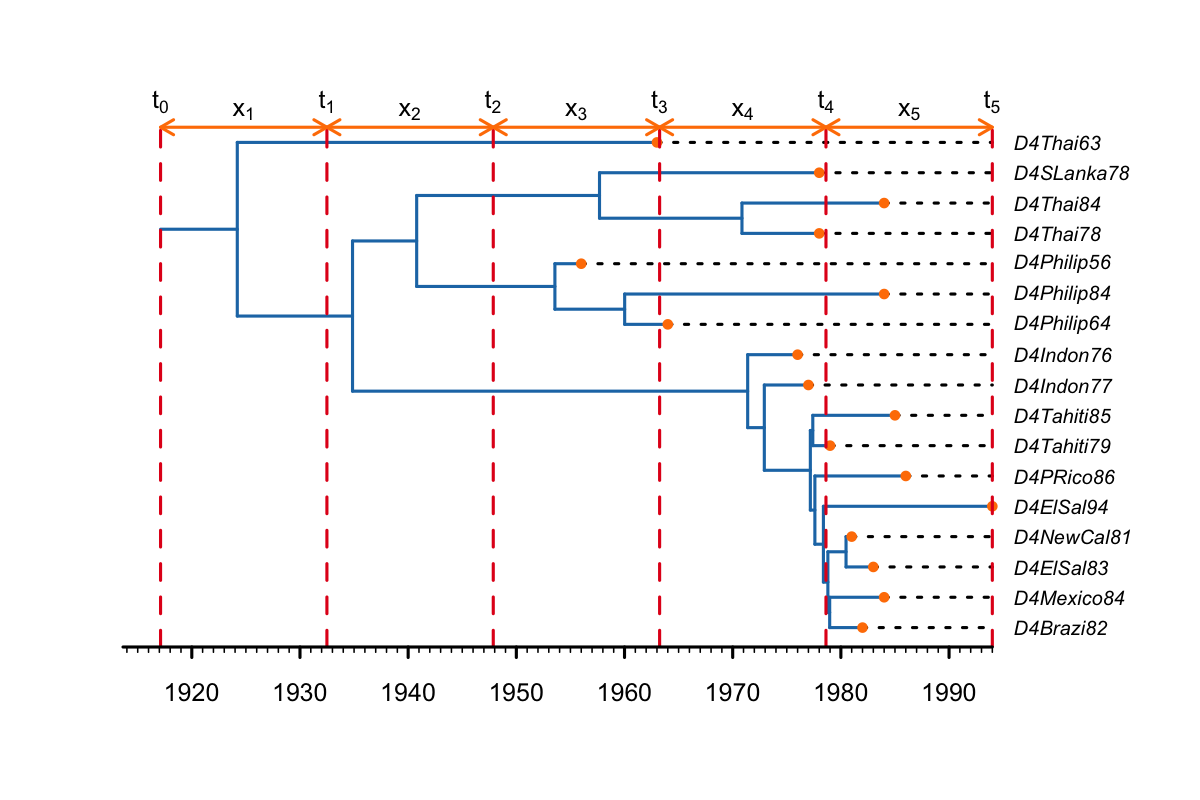
For most applications this is sufficient. However, when the origin branch is longer than one of the intervals, a change-point time will be placed on the origin branch (as below). This is problematic, because there is no information to identify rate-changes on the origin branch. Therefore, in some cases, equidistant change-point times between the origin and the present can cause problems with convergence and result in poor estimates.
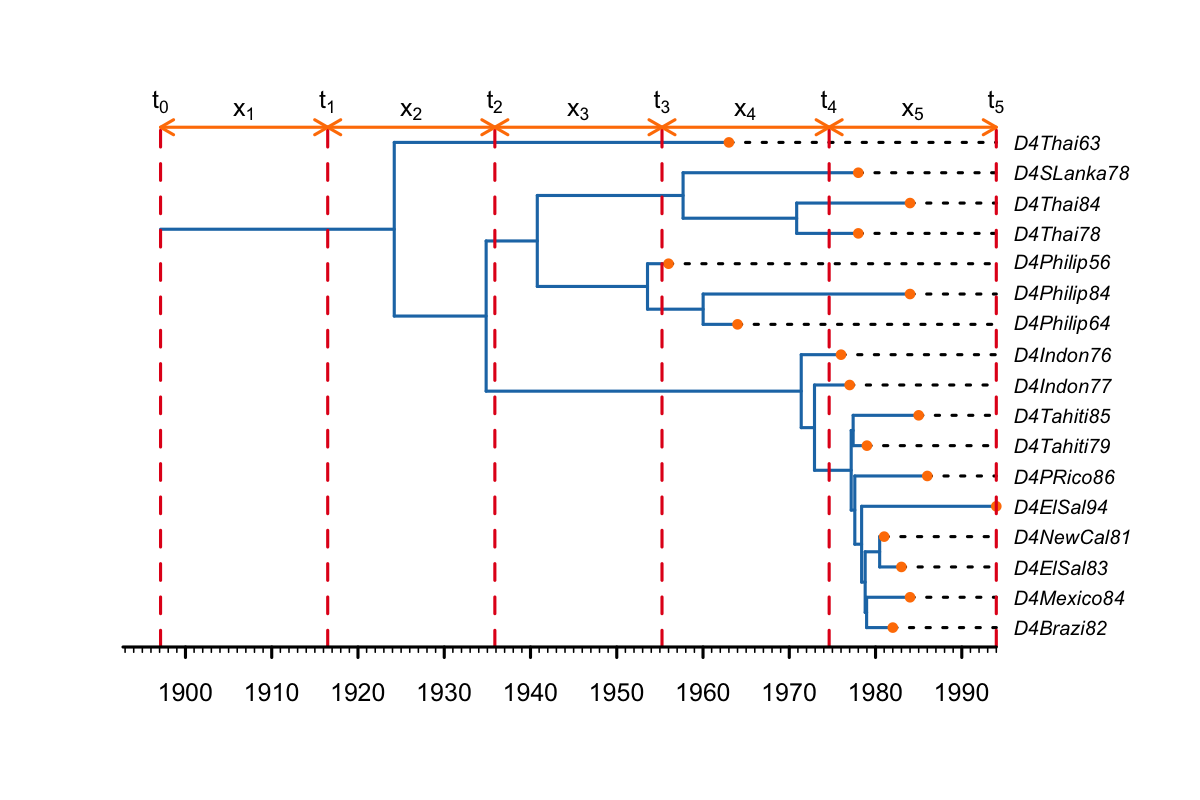
It is not possible to set equidistant change-point times between the tMRCA and the present using just BDSKY, without also fixing the tree.
The closest alternative is to condition the likelihood on the tMRCA of the tree, instead of the origin of the outbreak (by setting the option conditionOnRoot).
<distribution id="BirthDeathSkySerial"
spec="beast.evolution.speciation.BirthDeathSkylineModel"
tree="@Tree"
reproductiveNumber="@reproductiveNumber"
becomeUninfectiousRate="@becomeUninfectiousRate"
samplingProportion="@samplingProportion"
conditionOnRoot="true"/>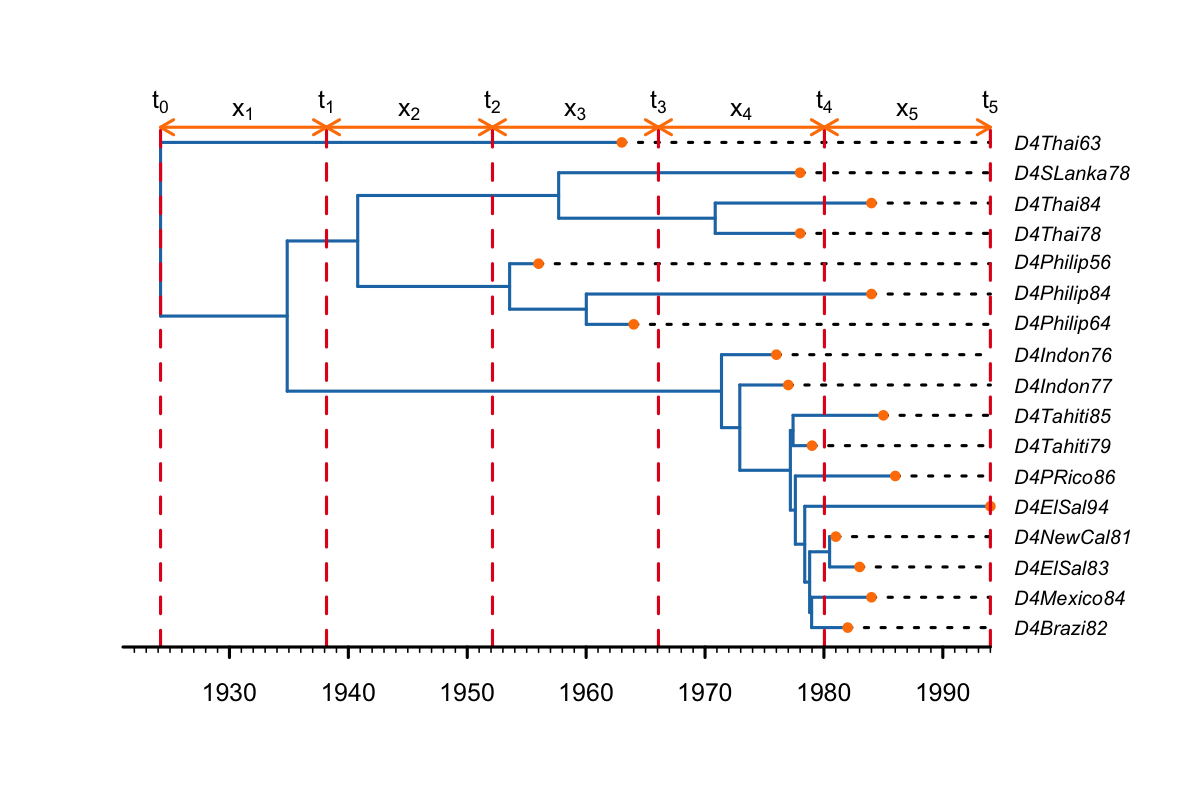
Note that in this case the origin parameter is not estimated at all (in fact it is not part of the model).
Example XML: dengue4_bdsky_equidistant5_treeslicer.xml
With TreeSlicer, we change birthRateChangeTimes to a TreeSlicer object, and explicitly tell it to place the change-point times between the tMRCA and the present.
<distribution id="BirthDeathSkySerial"
spec="beast.evolution.speciation.BirthDeathSkylineModel"
tree="@Tree"
reproductiveNumber="@reproductiveNumber"
becomeUninfectiousRate="@becomeUninfectiousRate"
samplingProportion="@samplingProportion"
origin="@origin">
<birthRateChangeTimes spec="TreeSlicer" id="ReTreeSlice" tree="@Tree"
dimension="5" to="tmrca" inclusive="false"/>
<reverseTimeArrays spec="BooleanParameter" value="true false false false false false"/>
</distribution>
We also need to indicate to BDSKY that the reproductive number (or birth-rate) change-point times should be reversed, meaning that change-point times are measured as time between the present and the change-point time (heights on the tree), instead of between the origin and the change-point time (default behaviour for BDSKY). We do this using the reverseTimeArrays array. Reproductive number (or birth-rate) change-point times are the first in the array (see BDSKY code/documentation), which is why the first element is set to true.
If we had set inclusive to true in the TreeSlicer object, there would be a change-point time on the tMRCA, as shown below. Convergence is generally better when inclusive is set to false (and the tMRCA is part of the oldest slice).
We can also log the TreeSlicer object,
<log idref="ReTreeSlice"/>which will record the time between the present and the reproductive number change-point times (heights on the tree) in the log file. Alternatively, we can directly log the dates of the change-point times with TreeSliceDateLogger,
<log spec="TreeSliceDateLogger" treeSlice="@ReTreeSlice"/>These times are needed to calculate marginal posterior distribution of the reproductive number at specific times, since the change-point times change with the tMRCA, which is one of the model parameters. (Actually, since change-point times are equally-spaced between the tRMCA and the present, only the tMRCA is needed to calculate the marginal posterior distribution).
Louis du Plessis, 2018