-
Notifications
You must be signed in to change notification settings - Fork 0
Efficient data analysis
Here we'll cover two packages that are useful for analyzing data efficiently and cleanly, plyr and dplyr. The two are very similar, however dplyr is an updated version of plyr just for data frames. We'll focus mainly on dplyr, with some comments on plyr at the end.
library(dplyr)Often what you'll want to do with a data frame is not necessarily move through it row by row but group by group. In other words, you want to
- split it apart into groups according to values in one of the columns,
- apply an action or multiple actions to the group, then
- combine the result of your actions back into a single table.
Recall the practice problem: Get the minimum value, maximum value, range of values, and median value of petal widths for each species.
You could do this in a for loop, where you subset iris by species and then run your actions on the subset. An example would be something like this (but there are zillion ways to accomplish the same task):
spp <- levels(iris$Species)
# create an empty vector to seed the data frame that will store values
spp.vals <- c()
for(i in 1:length(spp)){
# subset by species name and isolate Petal.Width
species.petal.w <- subset(iris, Species == spp[i])$Petal.Width
# add a new row to spp.vals with all the sought after values
spp.vals <- rbind(spp.vals, c(Species = spp[i],
min = min(species.petal.w),
max = max(species.petal.w),
range = diff(range(species.petal.w)),
median = median(species.petal.w)))
# move on to next species
}
spp.valsNotice again here we have to write the values that we want to keep to an outside variable, since the next iteration of the for loop will rewrite all the variables in the curly brackets.
The dplyr package provides functions that streamline this process using intuitive function names. For example, the above for loop can be rewritten using dplyr functions group_by() and summarise(). Notice you don't have to use dollar signs or anything for it to recognize what column you are trying to refer to.
# identify groups
iris.grouped <- group_by(iris, Species)
# create summary columns with an entry from each group
spp.vals <- summarise(iris.grouped,
min = min(Petal.Width),
max = max(Petal.Width),
range = diff(range(Petal.Width)),
median = median(Petal.Width))
# if you're done doing all the splitting, you can convert it
# from a "tibble" back into a plain old data frame
spp.vals <- as.data.frame(spp.vals)
spp.valsIf we wanted something more complex, we can write our own function! We apply this to our groups using the do() function.
The . symbol is a stand-in for your subsetted data frame ("group").
summariseIris <- function(df){
# this function returns a named data frame including the following values
data.frame(min = min(df$Petal.Width),
max = max(df$Petal.Width),
range = diff(range(df$Petal.Width)),
median = median(df$Petal.Width))
}
# check to make sure our function works with a data frame we know
summariseIris(iris)
spp.vals <- do(iris.grouped, summariseIris(.))
spp.vals <- as.data.frame(spp.vals)
spp.valsThe line below loads a dataset of fossil occurrences of whales.
Using any approach you feel comfortable using, tell me how many unique genera have been found in each county.
Store your counts in a data frame called
county.diversitywhere one column has your county ("county") and another column has your genus count ("county_div").whales <- read.csv("https://paleobiodb.org/data1.2/occs/list.txt?base_name=Cetacea&interval=Miocene&show=loc,class")Bonus: Merge
county.diversityintowhales. Now you have a column that tells you how many genera shared a county with each occurrence!
The package dplyr also introduces the ability to chain functions together using the %>% operator. Essentially, the output of one line gets used as the first argument of the next line. You can chain together any functions, not just ones included in dplyr.
This approach encourages you to write functions in the sequence you expect them to be applied (as opposed to being nested). It also helps reduce the number of variables you make.
# simple example 1
sqrt(c(4, 16))
# equivalent to:
c(4, 16) %>%
sqrt
# simple example 2
sum(sqrt(c(4, 16))) # sqrt happens before sum, but here is written after it
# equivalent to:
c(4, 16) %>%
sqrt %>%
sum# instead of running all your actions on separate lines:
mtcars
mtcars1 <- group_by(mtcars, cyl, am)
mtcars2 <- select(mtcars1, mpg, cyl, wt, am)
mtcars3 <- summarise(mtcars2, avgmpg = mean(mpg), avgwt = mean(wt))
mtcars4 <- filter(mtcars3, avgmpg > 20)
mtcars.final <- as.data.frame(mtcars4)
mtcars.final
# you can create a pipeline:
mtcars.final <- mtcars %>%
group_by(cyl, am) %>%
select(mpg, cyl, wt, am) %>%
summarise(avgmpg = mean(mpg), avgwt = mean(wt)) %>%
filter(avgmpg > 20) %>%
as.data.frame
mtcars.finalRewrite the fossil whale example into one chain of functions.
So, I mentioned that dplyr is a version of plyr specifically geared towards making analyses with data frames go smoother.
plyr, however, has a few functions that split more than just data frames and combine into more than just data frames. You have more control.
install.packages("plyr")
library(plyr)ldply() runs a function (custom or otherwise) over each element in a list and returns a data frame. This is very similar to lapply(), but you designated that the output must be in a data frame structure.
stuff.named <- list(numbers = c(66:101), word = "second", dat = mtcars)
# find the length of each element in the list called stuff
lapply(stuff.named, FUN = length)
# return it as a data frame instead
ldply(stuff.named, length)On the flip side, dlply() takes a data frame, split-apply-combines it, then returns a list. The syntax for splitting data frames apart in plyr is slightly different than in dplyr - you use the argument .(column_name) to designate what column has the groups in it.
# run a regression on Sepal.Length and Sepal.Width for each Species of iris separately
sepal.model <- function(sp){
return(lm(Sepal.Length ~ Sepal.Width, data = sp))
}
# test that your function works on something you know
sepal.model(iris)
model.per.sp <- dlply(iris, .(Species), sepal.model)
model.per.sp
str(model.per.sp)Make a data frame that has all the coefficients from the per-species regressions above.
Use whatever approach you'd like:
- apply functions
- dplyr functions
- plyr functions
- for loop Figure out what works for you!
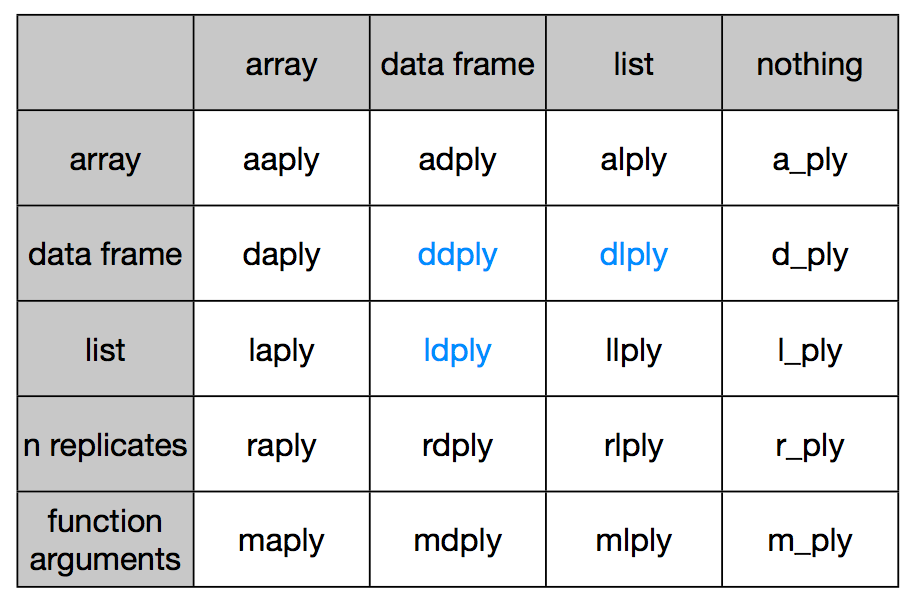
A good overview of what plyr functions can do can be found here.
Vertical says what goes in, horizontal says what comes out.
Day 1 or 2
- Basics of R
- Variables and objects
- Functions and packages
- Conditionals and logical operators
- Practice
- Recap
Day 3
Day 4
Day 5
Extra