This branch uses SLiM 3.6 (https://messerlab.org/slim/) to model how the mutational constraint on genes can influence polygenic adaptation to a new fitness optimum under a quantitative additive model for a single trait.
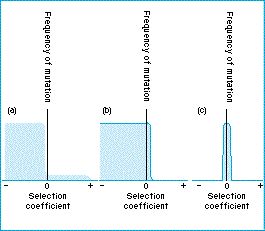
Each gene has a specified level of constraint, which determines the ratios of mutation types that can occur at each site. This approximates the classic figures from neutral theory we have all seen:
Where each gene has an individual one of these figures, and they can be of differing shape depending on constraint. For example, highly constrained genes (such as those encoding histones) will extremely rarely mutate favourably, so their mutations are mostly deleterious (and greatly so), with fewer neutral mutations, and almost no beneficial mutations. Trait fitness is calculated based on additive phenotype effects from trait mutations based on Lande's (1976)2 Gaussian fitness model. These phenotype effects are sampled from a normal distribution, N(0, σ), where σ is a parameter varied across treatments.
Populations are challenged to adapt to a new phenotypic optimum from a burnt-in starting position. A variety of genetic parameters are adjusted including
- Population size
- Proportion of the genome under high, medium, and low constraint
- Genome-wide recombination rate
- The number of QTLs contributing to the trait
- The strength of selection
- The size of the phenotypic shift (initial distance to the optimum)
- Variance in additive phenotype effects
To explore this parameter space, I use Latin hypercube sampling, courtesy of the R packages DoE.wrapper and
LHS. I then run each parameter combination 48 times with seeds generated from my own Mersenne Twister based
64-bit seed generator, seedgenerator, which can be found in ./src/Tools/SeedGenerator/.
1 Ridley, 2003, Evolution, Blackwell Publishing, https://www.blackwellpublishing.com/ridley/images/neutral_theory.jpg
2 Lande, 1976, Natural Selection and Random Genetic Drift in Phenotypic Evolution, Evolution 30:314-334