-
Notifications
You must be signed in to change notification settings - Fork 3
Home
Gut Analysis Toolbox enables semi-automated analysis of the neuronal and glial distribution within the gut wall (enteric nervous system). It uses a combination of FIJI macros, StarDist and deepImageJ under the hood to do the heavy lifting with automated segmentation. The deep learning models supplied with GAT enable automated segmentation of:
- enteric neuron soma (StarDist)
- enteric neuronal subtypes (StarDist)
- ganglia (deepImageJ)
The models have been trained using the easy to use notebooks from ZeroCostDL4Mic. The training data including the notebooks will be made available soon. The models will also be deposited on the BioImage Model Zoo.
Click on the sidebar to navigate through the WIKI.
Disclaimer: It is highly recommended to verify the output from the model and not to use it blindly. We have put a lot of effort into making sure the model works across a range of 2D images, but it is very likely that you may have to tune parameters by testing on your images. In some cases, you may need to train a new model or can contribute data for training.
You can also watch this as video tutorials on youtube by clicking on the image below.
Over time, we will be introducing features such as multiplex registration and analysis, calcium imaging analysis and analysis of other cell types/image modalities related to gut research. Feel free to post an Issue.
Download FIJI from this link: https://imagej.net/software/fiji/downloads and unzip it on your computer.
- To configure GAT, you will need to install a few plugins first

- You will get a pop up window that will check for updates:

- Once its done, the ImageJ Updater box will appear. Click Manage update sites on the bottom left and a window pops up:

- To install a plugin, tick on the corresponding boxes.

-
For installing GAT, tick on the following boxes:
- BIG-EPFL
- CSBDeep
- clij
- clij2
- DeepImageJ
- IJBP-Plugins (MorphoLibJ)
- StarDist
-
You will also need to manually add the GAT update site. Click on “Add update site”

- A new row will appear with empty boxes under URL column.

- Enter the following URL in the box: https://sites.imagej.net/GutAnalysisToolbox/

- Double click on “New” which is under the “Name” column and enter “GAT”.

-
Hit Enter and then click “Close”.
-
All the dependencies and scripts from the update sites selected above will appear in the ImageJ Updater.

- Click “Apply changes” and once its finished, restart FIJI.



In the newest version of GAT, the model files will be downloaded and installed automatically from the update site. If it is not working, please update ImageJ by going to Help -> Update ImageJ. Once updated, restart Fiji. If you already installed GAT, untick GAT update site and try the GAT installation process again.
- The models for neuron and ganglia segmentation are within
Fiji.app/modelsfolder. They contains models for:- Enteric neuron model: 2D_enteric_neuron_v4_1.zip
- Enteric neuron subtype model: 2D_enteric_neuron_subtype_v4_1.zip
- Ganglia model: 2D_enteric_ganglia_v2.bioimage.io.model
The download may take a while as the model files are large. After all the installation and configuration, when you restart FIJI, you should see the GAT menu appear on FIJI.

For a Mac, you can open the FIJI folder by right clicking on the folder:

GAT will be updated with features every now and then especially if any bugs have been reported. Usually FIJI may let you know if there are any updates. If not, you can manually update this by going to Help -> Update
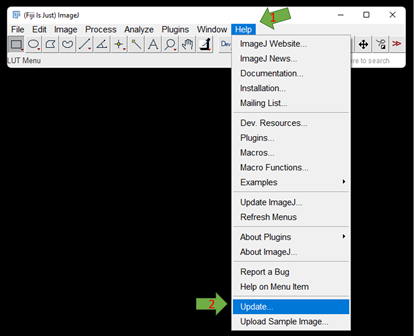
If there are any updates, it will appear in the Updater window:

Click Apply changes. The updates will be downloaded. Once its done, make sure you restart Fiji. The newest version of GAT should be available.
